Seurat单细胞处理流程之六:hdWGCNA分析
rm(list = ls())
setwd("/mnt/DEV_8T/zhaozm/seurat全流程/hdWGCNA/")
##禁止转化为因子
options(stringsAsFactors = FALSE)
## 设置保存的目录
## 数据目录
data_dir <- "/mnt/DEV_8T/zhaozm/seurat全流程/hdWGCNA/data"
if (!dir.exists(data_dir)) {
dir.create(data_dir, recursive = TRUE)
}
## 图片目录
img_dir <- "/mnt/DEV_8T/zhaozm/seurat全流程/hdWGCNA/img"
if (!dir.exists(img_dir)) {
dir.create(img_dir, recursive = TRUE)
}
library(Seurat)
library(tidyverse)
library(cowplot)
library(patchwork)
library(WGCNA)
library(hdWGCNA)
library(igraph)
library(Matrix)
library(tidyverse)
library(readxl)
library(ggh4x)
library(ggstar)
library(ggnewscale)
library(ggfun)
载入需要的程序包:SeuratObject
载入需要的程序包:sp
载入程序包:‘SeuratObject’
The following objects are masked from ‘package:base’:
intersect, t
载入程序包:‘cowplot’
The following object is masked from ‘package:lubridate’:
stamp
载入程序包:‘patchwork’
The following object is masked from ‘package:cowplot’:
align_plots
载入需要的程序包:dynamicTreeCut
载入需要的程序包:fastcluster
载入程序包:‘fastcluster’
The following object is masked from ‘package:stats’:
hclust
载入程序包:‘WGCNA’
The following object is masked from ‘package:stats’:
cor
载入需要的程序包:harmony
载入需要的程序包:Rcpp
载入需要的程序包:ggrepel
载入需要的程序包:igraph
载入程序包:‘igraph’
The following objects are masked from ‘package:lubridate’:
%--%, union
The following objects are masked from ‘package:dplyr’:
as_data_frame, groups, union
The following objects are masked from ‘package:purrr’:
compose, simplify
The following object is masked from ‘package:tidyr’:
crossing
The following object is masked from ‘package:tibble’:
as_data_frame
The following object is masked from ‘package:Seurat’:
components
The following objects are masked from ‘package:stats’:
decompose, spectrum
The following object is masked from ‘package:base’:
union
载入需要的程序包:ggraph
载入程序包:‘ggraph’
The following object is masked from ‘package:sp’:
geometry
载入需要的程序包:tidygraph
载入程序包:‘tidygraph’
The following object is masked from ‘package:igraph’:
groups
The following object is masked from ‘package:stats’:
filter
载入需要的程序包:UCell
载入需要的程序包:GeneOverlap
载入需要的程序包:GenomicRanges
载入需要的程序包:stats4
载入需要的程序包:BiocGenerics
载入程序包:‘BiocGenerics’
The following objects are masked from ‘package:igraph’:
normalize, path, union
The following objects are masked from ‘package:lubridate’:
intersect, setdiff, union
The following objects are masked from ‘package:dplyr’:
combine, intersect, setdiff, union
The following object is masked from ‘package:SeuratObject’:
intersect
The following objects are masked from ‘package:stats’:
IQR, mad, sd, var, xtabs
The following objects are masked from ‘package:base’:
anyDuplicated, aperm, append, as.data.frame, basename, cbind,
colnames, dirname, do.call, duplicated, eval, evalq, Filter, Find,
get, grep, grepl, intersect, is.unsorted, lapply, Map, mapply,
match, mget, order, paste, pmax, pmax.int, pmin, pmin.int,
Position, rank, rbind, Reduce, rownames, sapply, saveRDS, setdiff,
table, tapply, union, unique, unsplit, which.max, which.min
载入需要的程序包:S4Vectors
载入程序包:‘S4Vectors’
The following objects are masked from ‘package:tidygraph’:
active, rename
The following objects are masked from ‘package:lubridate’:
second, second<-
The following objects are masked from ‘package:dplyr’:
first, rename
The following object is masked from ‘package:tidyr’:
expand
The following object is masked from ‘package:utils’:
findMatches
The following objects are masked from ‘package:base’:
expand.grid, I, unname
载入需要的程序包:IRanges
载入程序包:‘IRanges’
The following object is masked from ‘package:tidygraph’:
slice
The following object is masked from ‘package:lubridate’:
%within%
The following objects are masked from ‘package:dplyr’:
collapse, desc, slice
The following object is masked from ‘package:purrr’:
reduce
The following object is masked from ‘package:sp’:
%over%
载入需要的程序包:GenomeInfoDb
Warning message:
“replacing previous import ‘GenomicRanges::intersect’ by ‘SeuratObject::intersect’ when loading ‘hdWGCNA’”
Warning message:
“replacing previous import ‘GenomicRanges::union’ by ‘dplyr::union’ when loading ‘hdWGCNA’”
Warning message:
“replacing previous import ‘GenomicRanges::setdiff’ by ‘dplyr::setdiff’ when loading ‘hdWGCNA’”
Warning message:
“replacing previous import ‘dplyr::as_data_frame’ by ‘igraph::as_data_frame’ when loading ‘hdWGCNA’”
Warning message:
“replacing previous import ‘Seurat::components’ by ‘igraph::components’ when loading ‘hdWGCNA’”
Warning message:
“replacing previous import ‘dplyr::groups’ by ‘igraph::groups’ when loading ‘hdWGCNA’”
Warning message:
“replacing previous import ‘dplyr::union’ by ‘igraph::union’ when loading ‘hdWGCNA’”
Warning message:
“replacing previous import ‘GenomicRanges::subtract’ by ‘magrittr::subtract’ when loading ‘hdWGCNA’”
Warning message:
“replacing previous import ‘Matrix::as.matrix’ by ‘proxy::as.matrix’ when loading ‘hdWGCNA’”
Warning message:
“replacing previous import ‘igraph::groups’ by ‘tidygraph::groups’ when loading ‘hdWGCNA’”
载入程序包:‘Matrix’
The following object is masked from ‘package:S4Vectors’:
expand
The following objects are masked from ‘package:tidyr’:
expand, pack, unpack
ggfun v0.1.8 Learn more at https://yulab-smu.top/
载入程序包:‘ggfun’
The following objects are masked from ‘package:cowplot’:
get_legend, theme_nothing
## 针对某一类细胞亚群做需要使用这个细胞亚群的高变基因,而不是全部的
# 载入数据
seurat_obj <- read_rds(file = "../差异富集/data/cd8注释后.rds")
seurat_obj <- subset(seurat_obj, subset = celltype == "Tex_CD8")
#寻找高变基因
seurat_obj <- FindVariableFeatures(seurat_obj,
selection.method = "vst",
nfeatures = 2000)
# 标准化
seurat_obj <- NormalizeData(seurat_obj, normalization.method = "LogNormalize", scale.factor = 10000)
## 挑选基因
seurat_obj <- SetupForWGCNA(
seurat_obj,
features = VariableFeatures(seurat_obj),
wgcna_name = "tutorial"
)
length(seurat_obj@misc$tutorial$wgcna_genes)
Finding variable features for layer counts
Normalizing layer: counts
2000
## 构建matacell
## k值根据实际情况再做调整
seurat_obj <- MetacellsByGroups(
seurat_obj = seurat_obj,
group.by = "group",
k = 10,
max_shared = 15,
min_cells = 20,
ident.group = 'group'
)
# 标准化metacell矩阵
seurat_obj <- NormalizeMetacells(seurat_obj)
Normalizing layer: counts
## 共表达网络分析
## 这一步将指定将用于网络分析的表达矩阵
seurat_obj <- SetDatExpr(
seurat_obj,
group_name = "Resistant", # 细胞名称
group.by='group', #
assay = 'RNA',
slot = 'data'
)
..Excluding 157 genes from the calculation due to too many missing samples or zero variance.
## 挑选软阈值
seurat_obj <- TestSoftPowers(
seurat_obj,
networkType = 'unsigned'
)
pickSoftThreshold: will use block size 1843.
pickSoftThreshold: calculating connectivity for given powers...
..working on genes 1 through 1843 of 1843
Warning message:
“executing %dopar% sequentially: no parallel backend registered”
Warning message in (function (x, y = NULL, robustX = TRUE, robustY = TRUE, use = "all.obs", :
“bicor: zero MAD in variable 'x'. Pearson correlation was used for individual columns with zero (or missing) MAD.”
Warning message in (function (x, y = NULL, robustX = TRUE, robustY = TRUE, use = "all.obs", :
“bicor: zero MAD in variable 'y'. Pearson correlation was used for individual columns with zero (or missing) MAD.”
Power SFT.R.sq slope truncated.R.sq mean.k. median.k. max.k.
1 1 0.0405 -0.302 0.9040 256.0000 2.35e+02 544.00
2 2 0.8280 -1.400 0.9670 64.0000 4.68e+01 249.00
3 3 0.9200 -1.580 0.9810 22.3000 1.20e+01 139.00
4 4 0.9120 -1.610 0.9680 9.6200 3.82e+00 86.40
5 5 0.9190 -1.590 0.9730 4.7900 1.44e+00 57.60
6 6 0.9000 -1.610 0.9630 2.6600 5.91e-01 40.20
7 7 0.9180 -1.590 0.9750 1.5900 2.51e-01 29.10
8 8 0.9220 -1.580 0.9650 1.0200 1.17e-01 21.60
9 9 0.9260 -1.570 0.9720 0.6900 5.57e-02 16.40
10 10 0.9310 -1.550 0.9880 0.4880 2.71e-02 12.60
11 12 0.8890 -1.490 0.9350 0.2770 6.71e-03 7.76
12 14 0.9140 -1.280 0.8910 0.1800 1.91e-03 5.06
13 16 0.8120 -1.400 0.7610 0.1320 5.50e-04 4.93
14 18 0.2530 -2.240 0.0441 0.1050 1.55e-04 4.83
15 20 0.2520 -2.130 0.0437 0.0893 4.54e-05 4.76
16 22 0.2510 -2.040 0.0431 0.0796 1.43e-05 4.69
17 24 0.2510 -1.970 0.0423 0.0732 4.71e-06 4.64
18 26 0.2650 -1.960 0.0773 0.0689 1.52e-06 4.60
19 28 0.2610 -1.910 0.0685 0.0658 4.89e-07 4.56
20 30 0.2590 -1.830 0.0679 0.0635 1.57e-07 4.52
## 使用ggplot2对图片的文字大小进行调整
plot_list <- PlotSoftPowers(seurat_obj,selected_power = 9)
# assemble with patchwork
# wrap_plots(plot_list, ncol=2)
## ggplot2调整
plot_list <- lapply(plot_list, function(p) {
layers <- lapply(p$layers, function(layer) {
if ("GeomText" %in% class(layer$geom)) {
layer$aes_params$size <- 6 # 这里调整字体大小
}
return(layer)
})
p$layers <- layers
return(p)
})
plot_list <- lapply(plot_list, function(p) {
p +
theme(text = element_text(size = 15),
axis.text = element_text(size = 12), # x/y 轴刻度字体大小
axis.title = element_text(size = 14), # x/y 轴标题字体大小
plot.title = element_text(size = 16, face = "bold"), # 标题加粗
legend.text = element_text(size = 12), # 图例文本字体大小
legend.title = element_text(size = 14) # 图例标题字体大小
)
})
# 然后使用 wrap_plots 组合图表
combined_plot <- wrap_plots(plot_list, ncol = 2)
# 打印组合图
print(combined_plot)
## 输出为pdf的时候长一些就行了
Power SFT.R.sq slope truncated.R.sq mean.k. median.k. max.k.
1 1 0.04047244 -0.3015199 0.9035691 255.756815 235.082186 543.51348
2 2 0.82794687 -1.3986747 0.9666878 64.000551 46.827492 249.29709
3 3 0.91995250 -1.5767948 0.9806154 22.308734 11.950025 138.96099
4 4 0.91219705 -1.6127395 0.9682106 9.616347 3.818667 86.40203
5 5 0.91856220 -1.5940438 0.9733032 4.794536 1.439149 57.57644
6 6 0.90018617 -1.6131170 0.9626550 2.655953 0.590680 40.23285
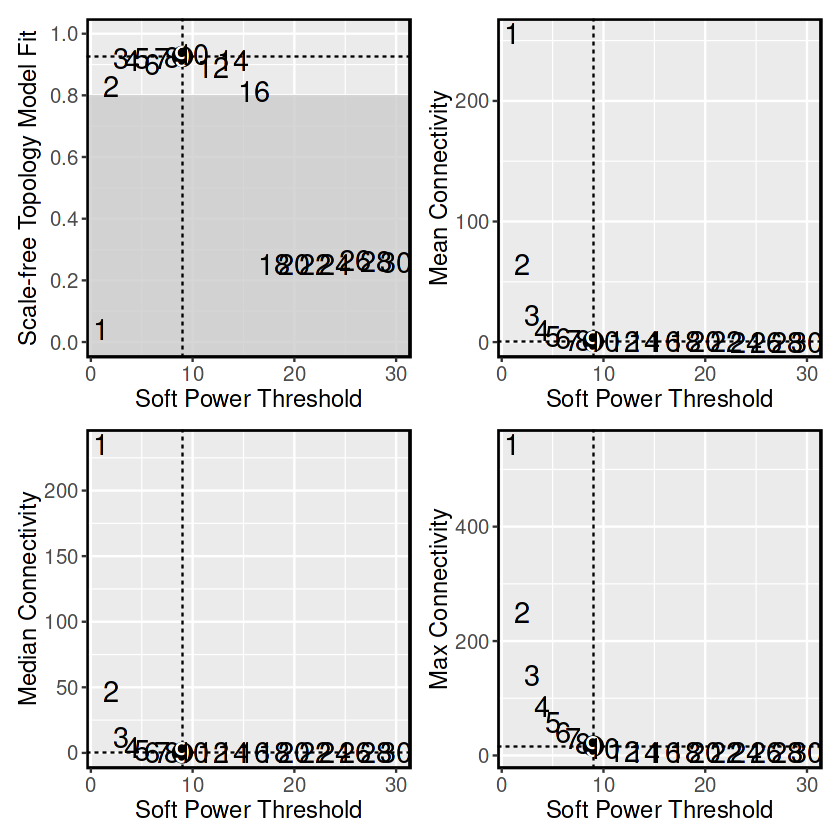
## 可以使用 GetPowerTable 函数访问以进行进一步检查:
power_table <- GetPowerTable(seurat_obj)
head(power_table)
| Power | SFT.R.sq | slope | truncated.R.sq | mean.k. | median.k. | max.k. | |
|---|---|---|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | |
| 1 | 1 | 0.04047244 | -0.3015199 | 0.9035691 | 255.756815 | 235.082186 | 543.51348 |
| 2 | 2 | 0.82794687 | -1.3986747 | 0.9666878 | 64.000551 | 46.827492 | 249.29709 |
| 3 | 3 | 0.91995250 | -1.5767948 | 0.9806154 | 22.308734 | 11.950025 | 138.96099 |
| 4 | 4 | 0.91219705 | -1.6127395 | 0.9682106 | 9.616347 | 3.818667 | 86.40203 |
| 5 | 5 | 0.91856220 | -1.5940438 | 0.9733032 | 4.794536 | 1.439149 | 57.57644 |
| 6 | 6 | 0.90018617 | -1.6131170 | 0.9626550 | 2.655953 | 0.590680 | 40.23285 |
## 使用ConstructNetwork函数构建共表达网络
## 根据上一步的结果,分析的细胞为Exhausted CD8 T cell
seurat_obj <- ConstructNetwork(
seurat_obj, soft_power=9,
setDatExpr=FALSE,
tom_name = 'Tex_CD8'
)
Calculating consensus modules and module eigengenes block-wise from all genes
Calculating topological overlaps block-wise from all genes
Flagging genes and samples with too many missing values...
..step 1
TOM calculation: adjacency..
..will not use multithreading.
Fraction of slow calculations: 0.000000
..connectivity..
..matrix multiplication (system BLAS)..
..normalization..
..done.
..Working on block 1 .
..Working on block 1 .
..merging consensus modules that are too close..
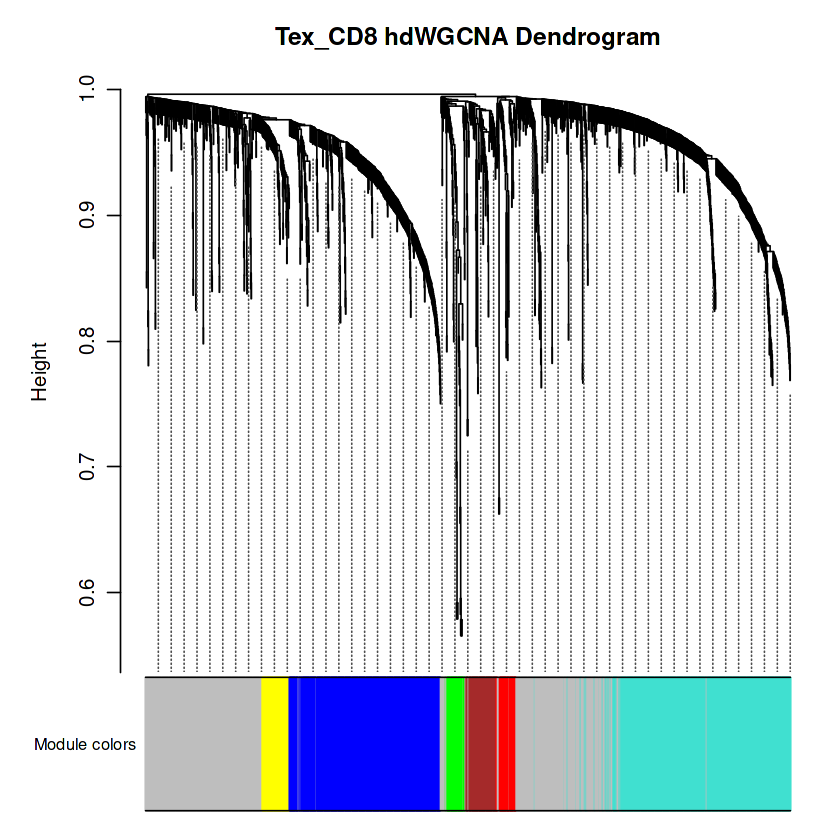
## 可视化
PlotDendrogram(seurat_obj, main='Tex_CD8 hdWGCNA Dendrogram')
## 灰色模块占比不能太多,否则大部分的基因没有意义
## 保存相关性矩阵
TOM <- GetTOM(seurat_obj)
TOM <- as.data.frame(TOM)
write.csv(TOM,file = "./data/cor_Tex_CD8.csv")
## 查看模块构成基因
seurat_obj@misc$tutorial$wgcna_modules %>% head
table(seurat_obj@misc$tutorial$wgcna_modules$module)
| gene_name | module | color | |
|---|---|---|---|
| <chr> | <fct> | <chr> | |
| GNLY | GNLY | turquoise | turquoise |
| HBB | HBB | blue | blue |
| CCL4L2 | CCL4L2 | turquoise | turquoise |
| CCL4 | CCL4 | turquoise | turquoise |
| HBA2 | HBA2 | grey | grey |
| HSPA6 | HSPA6 | turquoise | turquoise |
turquoise blue grey red green brown yellow
522 429 626 50 52 86 78
## 计算每个单细胞的模块基因
seurat_obj <- ScaleData(seurat_obj, features=VariableFeatures(seurat_obj))
seurat_obj <- ModuleEigengenes(
seurat_obj,
group.by.vars="group"
) #时间较长
# harmonized module eigengenes:
hMEs <- GetMEs(seurat_obj)
# module eigengenes:
MEs <- GetMEs(seurat_obj, harmonized=FALSE)
# 计算枢纽基因
seurat_obj <- ModuleConnectivity(
seurat_obj,
group.by = 'celltype', group_name = 'Tex_CD8'
)
Centering and scaling data matrix
Warning message:
“Different features in new layer data than already exists for scale.data”
[1] "turquoise"
Centering and scaling data matrix
Warning message:
“Keys should be one or more alphanumeric characters followed by an underscore, setting key from pcaturquoise to pcaturquoise_”
pcaturquoise_ 1
Positive: GNLY, ENTPD1, GZMB, KLRD1, GAPDH, LAYN, TNFRSF18, LAG3, TIGIT, PHLDA1
JUN, KRT86, GALNT2, HSPB1, CCL3, CD7, FAM3C, HAVCR2, ACP5, KIR2DL4
MYO1E, CTLA4, CAPG, TPI1, NKG7, ITGAE, PTMS, CSF1, PLPP1, PRF1
Negative: CLECL1, EIF1AY, RILPL2, MTFP1, CD27, LITAF, HIST1H2AG, H2AFX, HIST1H2AM, KLRC3
PDE4DIP, RND1, SEC14L2, ANKRD37, HIST1H3J, HNRNPLL, FOXO1, BRSK1, CFAP20, SSBP3
SMARCD2, HIST1H4B, PAXIP1, IER5L, B3GNT7, BRCA2, PRDX2, HSPA2, ZNF101, GIMAP1
pcaturquoise_ 2
Positive: KRT81, TNFRSF18, S100A4, MYL6, KRT86, FAM166B, PDLIM4, CD63, HTRA1, GAPDH
KLRC2, MYO1E, PLPP1, GZMA, AC068775.1, IL2RA, ENTPD1, CTSW, TMSB4X, SCX
TMSB10, BEX3, DNPH1, FAM3C, CAPG, PLEKHA1, TSPAN13, MTSS1, CD7, DBN1
Negative: HSP90AA1, HSPA1B, DNAJB1, HSPA1A, HSP90AB1, DNAJA1, HSPH1, HSPD1, DUSP1, HSPE1
ZFAND2A, CHORDC1, JUN, UBC, MRPL18, FKBP4, ATF3, TAGAP, DNAJB4, HSPA6
BAG3, DDIT4, HSPB1, RGS1, RGS2, DDIT3, DUSP4, ANKRD37, NR4A1, RPS19
pcaturquoise_ 3
Positive: TMSB4X, TMSB10, NKG7, ACTB, SH3BGRL3, RPS19, MYL6, COTL1, GZMA, CD27
IFITM2, MIF, IFI6, CXCL13, S100A4, GAPDH, RPLP0, FKBP1A, FABP5, C12orf75
LITAF, ISG15, DYNLL1, GMFG, HMGN2, CCL4, ACTG1, H2AFZ, CCL4L2, APOBEC3G
Negative: JUN, ENTPD1, GNLY, PIM1, TNFRSF18, MYO1E, GPR25, KLRB1, SOX4, SNX9
GNA15, DAPK2, VDR, KLRD1, MTSS1, PHACTR2, CSF1, HPSE, LAYN, TMIGD2
HPGD, IL18RAP, BCL2L11, NFKBIZ, ATP8B4, SPRY1, THBS1, RHOB, SQLE, GADD45G
pcaturquoise_ 4
Positive: KRT81, KRT86, CAPG, HOPX, SH3BGRL3, TMSB4X, CTSW, TMIGD2, TMSB10, PPP1R14B
S100A4, FAM166B, DYNLL1, HSPE1, MYL6, ACTB, MYO7A, CD63, RPS19, KLRD1
CXXC5, CCL20, CLNK, ALOX5AP, HSPA1A, NUDT14, KLRC2, KLRB1, RPLP0, HTRA1
Negative: CCL4L2, LYST, CCL3L1, DUSP5, CD27, LAG3, VCAM1, CCL4, TNFRSF9, PRDM1
CXCL13, MX1, HAVCR2, SNX9, PDCD1, PRF1, DUSP4, CTLA4, SLA, TNIP3
RHOH, ZBTB32, HNRNPLL, NAB1, DUSP16, TGIF1, CHST12, TIGIT, CBLB, IPCEF1
pcaturquoise_ 5
Positive: GZMB, SLA2, PRF1, KIR2DL4, NKG7, CXCR6, CLEC2B, FASLG, CLIC3, GNLY
ABI3, GPR25, HMGB2, GABARAPL1, ACP5, ID2, ITGAE, WIPF3, FKBP4, GALNT2
POLR1E, CD7, BCL2L11, IFITM2, C12orf75, HAVCR2, TXNIP, RALA, GZMA, VCAM1
Negative: DUSP1, GNG4, GEM, PRDX2, CITED2, TMSB10, TNFRSF18, IFNG, FABP5, BEX3
PHACTR2, PDLIM4, ZFP36L1, RILPL2, RGS2, RPLP0, TOX2, ANKRD37, H2AFX, C16orf45
ACTG1, CNIH1, TNFRSF9, CEBPA, DNAJB1, PIP5K1B, HTRA1, CD27, IL26, FAM107B
Transposing data matrix
Initializing state using k-means centroids initialization
Harmony 1/10
Harmony 2/10
Harmony 3/10
Harmony 4/10
Harmony 5/10
Harmony 6/10
Harmony converged after 6 iterations
[1] "blue"
Centering and scaling data matrix
Warning message:
“Keys should be one or more alphanumeric characters followed by an underscore, setting key from pcablue to pcablue_”
pcablue_ 1
Positive: CD2, MAF, MT-ND2, USP51, APBB2, APP, SRRT, PHLDA2, CPEB4, CAVIN3
BIK, HLA-DPA1, PRDX4, IFI27, CTNND1, NCOA1, HHAT, CST3, RTN4R, TPSB2
CCL2, GPATCH4, NCAPH, RAB20, HLA-DRA, PCNX2, TMEM176A, FCHO2, MAP1LC3A, SLCO4C1
Negative: LMNA, IL7R, AHNAK, MYADM, SLC2A3, REL, FOSB, FOS, ANXA1, EZR
CXCR4, PTGER4, NR4A3, RORA, TNFAIP3, ELL2, CCR7, SORL1, VIM, PBXIP1
CDK17, BTG2, KLF3, ATP2B1, CD55, RANBP2, CEMIP2, MYH9, ZFP36, B4GALT1
pcablue_ 2
Positive: ANKRD28, PRNP, VIM, BHLHE40, XCL1, CD2, CD83, PERP, IVNS1ABP, IL12RB2
TIMP1, TMEM200A, NR3C1, RORA, GPR35, TNFSF14, TMEM123, GPR183, BIRC2, GLUD1
ARL6IP5, IL7R, XCL2, S100A9, PTGER4, IRF4, TNF, ITGB1, CHD4, ITM2A
Negative: DUSP2, GZMK, YPEL5, CXCR4, JUNB, SRSF5, TUBA4A, TC2N, CD74, IRF1
ZFP36L2, SRSF7, FAM177A1, ZFP36, ATP1B3, KLRG1, SRRT, RNF125, TGFB1, P2RY8
CHMP1B, SYAP1, FCMR, IDI1, BIK, SAT1, CEMIP2, MATK, MCUB, SLC2A3
pcablue_ 3
Positive: ZFP36L2, TUBA4A, SLC7A5, CXCR4, FAM177A1, TNFAIP3, AUTS2, PLEKHA2, CAMK4, SRSF7
EZR, PIK3R1, NR3C1, TNFSF14, IL21R, RANBP2, ARRDC3, NUP98, PPP1CB, VPS37B
REL, FBXO34, NFE2L2, GPR132, HIST1H1D, SATB1, PDE4A, SC5D, MYH9, SNRK
Negative: FOSB, FOS, HLA-DPA1, CD74, HLA-DRB1, MYADM, GZMK, HLA-DRA, NEU1, ARL6IP5
DDX3X, ITM2A, TAGLN2, PNP, LMNA, CD2, GLA, TUBA1A, ANXA1, GLUD1
CLDND1, SLC38A2, MCL1, RSRP1, DDX3Y, MT-ND2, ATP2B4, HSP90B1, IL27RA, CD69
pcablue_ 4
Positive: FOSB, FOS, CD69, ANXA1, MYADM, TUBA1A, ZFP36L2, IER2, GLUL, GADD45B
MATK, IVNS1ABP, NEU1, AUTS2, ZFP36, CAMK4, PLK3, BTG2, PNP, SCML4
MCL1, XCL1, IFRD1, XBP1, IRF1, MAFF, XCL2, SRSF5, SLC1A5, KLF10
Negative: CD28, CCR7, GPR183, GZMK, RNF19A, TCF7, FCMR, CLDND1, SYNE2, CD74
LTB, ANTXR2, ITM2A, GPCPD1, KLF3, RASA3, MAF, PBXIP1, CD81, P2RY8
S1PR1, ATM, APLP2, ICAM2, ZDBF2, CEMIP2, ITGA4, WDR7, SRRT, LSR
pcablue_ 5
Positive: HLA-DRB1, HLA-DRA, HLA-DPA1, CD74, ITM2C, VPS37B, BHLHE40, ITM2A, HAUS3, NFKB1
SLC7A5, TUBA4A, ANXA2, SELPLG, SERPINB9, FLNA, IRF4, IL21R, GZMK, SAMD9L
MATK, ARL6IP5, SRRT, PIK3R1, MYH9, G3BP2, UQCRC2, PLCG1, XCL2, TBC1D2B
Negative: IL7R, CCR7, FOS, FOSB, MT-ND2, LSR, TCF7, LMNA, ICAM2, MYADM
GPR183, NEU1, FCMR, JUNB, CHMP1B, DNAAF2, ANKRD28, CAMK1D, NR1D2, SLC38A2
MAF, IL12RB2, CD55, MS4A1, CD28, TUBA1A, PLXND1, MCUB, SPINT2, PBX4
Transposing data matrix
Initializing state using k-means centroids initialization
Harmony 1/10
Harmony 2/10
Harmony 3/10
Harmony 4/10
Harmony 5/10
Harmony converged after 5 iterations
Warning message:
“Key ‘harmony_’ taken, using ‘amiqp_’ instead”
[1] "grey"
Centering and scaling data matrix
Warning message:
“Keys should be one or more alphanumeric characters followed by an underscore, setting key from pcagrey to pcagrey_”
pcagrey_ 1
Positive: SRGAP3, LDLRAD4, CD9, IL2RB, ZEB2, CTSA, AHR, TNFSF10, BIRC3, RGS16
LAIR2, RAB3GAP1, OXNAD1, KLF6, BACH2, PIK3IP1, CCND2, ZNF267, JAML, AGPAT5
KLRC4, EIF2S3, SDC4, QPCT, IER5, ABCB1, DRAXIN, CDKN1A, BCL2, KIF5C
Negative: HIST1H4C, IFITM1, NR4A2, ENC1, FTL, ZNF331, BTG1, CCL5, TRAT1, HLA-DRB5
GZMH, TUBB4B, CD200R1, HLA-DQB1, AIF1, NMB, CDC34, MARCKSL1, TSHZ2, TNFSF8
JCHAIN, PLIN2, NCF1, ALG13, AREG, SOCS3, LYAR, UCP2, ARL4D, FTH1
pcagrey_ 2
Positive: LAIR2, CD9, SPP1, SRGAP3, PTGIS, S100A11, FOXP3, CADM1, MICAL2, FUT7
CD86, KIF5C, CORO1B, AGPAT5, TNFSF13B, SCPEP1, PDGFA, QPCT, CCND2, MYL9
LTA, CYSLTR1, CXCL8, IL2RB, FCER1G, DARS2, CTSC, IFITM3, HBA2, RSAD2
Negative: NR4A2, PPP1R15A, CSRNP1, ZNF331, KLF6, TUBB4B, RGCC, DDX5, TSC22D3, SERTAD1
KLF2, RASGEF1B, EGR1, CRTAM, ARL4A, FAM53C, TUBB2A, SOCS3, MAT2A, NAF1
SDCBP, LDHA, MARCKSL1, CRIP1, IER5, BTG1, AZIN1, HERPUD1, PIM3, ZC3H12A
pcagrey_ 3
Positive: PFKFB3, BTG1, ZNF331, HIST1H1E, HIST1H4C, ZBTB1, METRNL, PIK3IP1, SLC1A4, PTP4A1
FOSL2, SLFN5, LMNB1, SOCS3, ENC1, HIST1H1C, ASF1A, RHBDD2, S1PR4, UCP2
CDK16, NFKB2, EIF1AX, TP53INP1, GIMAP7, BCL3, STX11, PTK2B, TRAT1, TNFRSF4
Negative: PPP1R15A, CRIP1, EGR1, RASGEF1B, CCL5, SGK1, DDX5, TUBB4B, HLA-DQB1, CD9
ARL4A, KLF6, HLA-DRB5, SERTAD1, ARL4D, ID1, MLF1, ARG2, CTSA, GZMH
TSC22D3, LAIR2, FTL, CHAC1, S100A11, RBKS, GSTM3, RGS16, SPP1, EDARADD
pcagrey_ 4
Positive: TNFRSF4, CADM1, TSHZ2, GK, NMB, FBXO32, TRAT1, ENC1, CORO1B, RIC3
CYSLTR1, PCSK1N, CHI3L2, IGFBP4, MICAL2, FBLN7, CD200R1, PECAM1, TESPA1, KLF2
BICDL1, STX11, MPP1, NUCB2, STMN1, FOXP3, SERTAD1, LAIR2, GNA12, SMAD1
Negative: CCL5, GZMH, BTG1, FOSL2, SCUBE1, SYTL2, EIF1AX, LDLRAD4, PFKFB3, ZEB2
GBP5, OXNAD1, OSM, HIST1H1C, PIK3R6, KLRC4, HIST1H4C, MXD1, TSC22D3, PRMT9
ETS2, TRAF4, ZNF250, IL2RB, ABTB2, ABCB1, NUGGC, PIK3AP1, GAB3, SOCS1
pcagrey_ 5
Positive: FTH1, BTG1, CD9, LDLRAD4, RGCC, HIST1H4C, PFKFB3, SLC16A3, CDKN1A, RGS16
PMEPA1, IRS2, KIF5C, ZNF250, LAIR2, LDHA, FOXP3, RASGRP2, CDC34, NAF1
MGLL, PRMT9, SPP1, TRAF4, ZNF331, PIK3R6, EPHA4, GCHFR, AREG, BNIP3
Negative: CTSC, GZMH, IFITM1, GBP5, STAT1, CCL5, GIMAP7, ICOS, DTX3L, OAS3
GBP4, PARP9, STX11, RSAD2, HLA-DQB1, GIMAP5, IRF7, KLF6, TESPA1, TANK
HERC5, SGK1, KLF9, LAX1, TNFSF10, SLFN5, EGR2, EDARADD, CCR1, SNX18
Transposing data matrix
Initializing state using k-means centroids initialization
Harmony 1/10
Harmony converged after 1 iterations
[1] "red"
Centering and scaling data matrix
Warning message in irlba(A = t(x = object), nv = npcs, ...):
“You're computing too large a percentage of total singular values, use a standard svd instead.”
Warning message in irlba(A = t(x = object), nv = npcs, ...):
“did not converge--results might be invalid!; try increasing work or maxit”
Warning message:
“Keys should be one or more alphanumeric characters followed by an underscore, setting key from pcared to pcared_”
Warning message:
“Requested number is larger than the number of available items (50). Setting to 50.”
Warning message:
“Requested number is larger than the number of available items (50). Setting to 50.”
Warning message:
“Requested number is larger than the number of available items (50). Setting to 50.”
Warning message:
“Requested number is larger than the number of available items (50). Setting to 50.”
Warning message:
“Requested number is larger than the number of available items (50). Setting to 50.”
pcared_ 1
Positive: ZNF683, S100A10, CASS4, LGALS1, FLT3LG, DDAH2, NXPH4, MEF2D, TIAM2, DCTN1
CDKN2B, CTH, HIST1H4F, KRT8, CRYBG2, CCND1, NKRF, KRT7, IFIT2, HIST2H2AA4
CP, LST1, KIFC1, CDKL5, SPECC1
Negative: MYLIP, HOOK2, HIST2H2BF, GIMAP4, CEP290, TENT5A, SLC35G1, CHRNA1, NAP1L3, ASPM
EGR3, SPINK1, FOXJ1, CLMN, HES4, VSIG1, MPP5, OAT, LACTB2, FANCD2
KLF11, RCAN2, MAFB, KLF5, ZNF703
pcared_ 2
Positive: DDAH2, S100A10, LGALS1, ZNF683, CDKN2B, KRT8, CLMN, CP, SPINK1, CDKL5
KIFC1, NAP1L3, VSIG1, IFIT2, MAFB, FOXJ1, CASS4, CRYBG2, LST1, HIST1H4F
KRT7, FLT3LG, KLF5, NXPH4, HIST2H2AA4
Negative: MYLIP, OAT, TENT5A, CEP290, MEF2D, GIMAP4, ZNF703, CTH, FANCD2, KLF11
LACTB2, HIST2H2BF, NKRF, HOOK2, TIAM2, HES4, RCAN2, ASPM, SLC35G1, CCND1
SPECC1, EGR3, CHRNA1, MPP5, DCTN1
pcared_ 3
Positive: LGALS1, GIMAP4, CEP290, CDKN2B, S100A10, TENT5A, LACTB2, CTH, DCTN1, TIAM2
HIST2H2BF, KLF11, DDAH2, ASPM, CCND1, HIST1H4F, FLT3LG, MEF2D, KLF5, MAFB
KRT7, SPECC1, EGR3, CP, VSIG1
Negative: NKRF, HES4, CASS4, NXPH4, HOOK2, IFIT2, SLC35G1, ZNF683, OAT, HIST2H2AA4
ZNF703, KIFC1, FANCD2, RCAN2, FOXJ1, CHRNA1, CRYBG2, CLMN, LST1, NAP1L3
CDKL5, MPP5, SPINK1, KRT8, MYLIP
pcared_ 4
Positive: RCAN2, LACTB2, MEF2D, ZNF703, CDKN2B, LGALS1, HOOK2, CCND1, KRT7, S100A10
NAP1L3, ZNF683, ASPM, EGR3, TIAM2, KLF5, SPECC1, LST1, CRYBG2, CASS4
CDKL5, VSIG1, MYLIP, CP, OAT
Negative: DDAH2, GIMAP4, FLT3LG, CEP290, DCTN1, NKRF, IFIT2, SLC35G1, TENT5A, KIFC1
HES4, HIST2H2AA4, HIST1H4F, CTH, NXPH4, FANCD2, FOXJ1, KRT8, SPINK1, MPP5
MAFB, CHRNA1, KLF11, CLMN, HIST2H2BF
pcared_ 5
Positive: CTH, KLF11, LACTB2, HOOK2, GIMAP4, ZNF703, FANCD2, DDAH2, S100A10, EGR3
OAT, HES4, ASPM, HIST1H4F, HIST2H2BF, HIST2H2AA4, KLF5, MPP5, IFIT2, FOXJ1
CHRNA1, LGALS1, LST1, SPECC1, CDKL5
Negative: CEP290, TIAM2, MYLIP, FLT3LG, RCAN2, CDKN2B, MEF2D, NKRF, SLC35G1, TENT5A
DCTN1, CRYBG2, CCND1, CASS4, KRT7, MAFB, NXPH4, CP, KRT8, NAP1L3
VSIG1, KIFC1, CLMN, SPINK1, ZNF683
Transposing data matrix
Initializing state using k-means centroids initialization
Harmony 1/10
Harmony 2/10
Harmony 3/10
Harmony converged after 3 iterations
Warning message:
“Key ‘harmony_’ taken, using ‘easkb_’ instead”
[1] "green"
Centering and scaling data matrix
Warning message in irlba(A = t(x = object), nv = npcs, ...):
“You're computing too large a percentage of total singular values, use a standard svd instead.”
Warning message in irlba(A = t(x = object), nv = npcs, ...):
“did not converge--results might be invalid!; try increasing work or maxit”
Warning message:
“Keys should be one or more alphanumeric characters followed by an underscore, setting key from pcagreen to pcagreen_”
Warning message:
“Requested number is larger than the number of available items (52). Setting to 52.”
Warning message:
“Requested number is larger than the number of available items (52). Setting to 52.”
Warning message:
“Requested number is larger than the number of available items (52). Setting to 52.”
Warning message:
“Requested number is larger than the number of available items (52). Setting to 52.”
Warning message:
“Requested number is larger than the number of available items (52). Setting to 52.”
pcagreen_ 1
Positive: UBE2S, KRT17, CXCR1, LINC00672, ESCO2, PDLIM1, CLDN10, ZBED3, MUC12, CAVIN1
HIST1H3B, PTPRG, FAM167B, ITPKA, PPP1R2C, SIX3, TUBB4A, MT3, SH3RF2, ABCA1
OVOL1, CD24, FHL2, PMCH, UBE2E2, HLA-DQA2
Negative: CAV1, TIMD4, MYB, IL10, TMEM155, SELL, PGM2L1, ARMH1, FAM184A, PRDM8
CD200, ELOVL4, MT1G, ATL1, CTHRC1, S100A2, RNF157, TCEA3, CD300A, WFDC2
PCLAF, MYC, BPGM, DUSP6, UBTD1, KLF4
pcagreen_ 2
Positive: TMEM155, PDLIM1, ELOVL4, ATL1, BPGM, RNF157, MT1G, UBE2S, TIMD4, FAM184A
TUBB4A, CD300A, HLA-DQA2, DUSP6, IL10, OVOL1, ZBED3, CLDN10, WFDC2, HIST1H3B
UBTD1, LINC00672, KRT17, CXCR1, PCLAF, S100A2
Negative: ARMH1, CD200, MUC12, CAV1, SELL, KLF4, PRDM8, PGM2L1, CD24, PPP1R2C
TCEA3, MYC, UBE2E2, CAVIN1, FAM167B, PMCH, MT3, SH3RF2, SIX3, ABCA1
PTPRG, ITPKA, MYB, FHL2, CTHRC1, ESCO2
pcagreen_ 3
Positive: UBE2S, MUC12, HLA-DQA2, CD300A, TUBB4A, ELOVL4, DUSP6, BPGM, PRDM8, ARMH1
S100A2, RNF157, FHL2, KLF4, SELL, MT1G, MT3, HIST1H3B, KRT17, FAM167B
ESCO2, SIX3, CD24, SH3RF2, UBTD1, ABCA1
Negative: ATL1, WFDC2, PCLAF, FAM184A, TIMD4, PDLIM1, IL10, TMEM155, MYC, CTHRC1
PGM2L1, MYB, LINC00672, CAV1, CD200, CAVIN1, PPP1R2C, ZBED3, CLDN10, PTPRG
TCEA3, ITPKA, OVOL1, PMCH, UBE2E2, CXCR1
pcagreen_ 4
Positive: CD300A, TIMD4, S100A2, DUSP6, ELOVL4, WFDC2, MT1G, IL10, FHL2, RNF157
MYB, PCLAF, UBTD1, HIST1H3B, LINC00672, KRT17, CTHRC1, ESCO2, CAVIN1, SH3RF2
PMCH, FAM167B, SIX3, TUBB4A, CXCR1, OVOL1
Negative: PDLIM1, ATL1, UBE2S, CD200, SELL, ARMH1, MUC12, PRDM8, FAM184A, TMEM155
TCEA3, KLF4, MYC, HLA-DQA2, BPGM, PGM2L1, CD24, ZBED3, MT3, CAV1
CLDN10, UBE2E2, ITPKA, PTPRG, PPP1R2C, ABCA1
pcagreen_ 5
Positive: PGM2L1, PRDM8, MUC12, FAM184A, CAV1, HLA-DQA2, MYB, UBE2S, DUSP6, MT1G
UBE2E2, CTHRC1, MT3, PTPRG, ITPKA, IL10, TCEA3, FAM167B, KLF4, SIX3
MYC, CAVIN1, SH3RF2, HIST1H3B, PMCH, CXCR1
Negative: CD200, ARMH1, BPGM, SELL, CD300A, TUBB4A, S100A2, PCLAF, PDLIM1, WFDC2
TMEM155, FHL2, RNF157, PPP1R2C, ATL1, UBTD1, LINC00672, TIMD4, KRT17, CLDN10
OVOL1, ELOVL4, CD24, ESCO2, ABCA1, ZBED3
Transposing data matrix
Initializing state using k-means centroids initialization
Harmony 1/10
Harmony 2/10
Harmony 3/10
Harmony converged after 3 iterations
[1] "brown"
Centering and scaling data matrix
Warning message in irlba(A = t(x = object), nv = npcs, ...):
“You're computing too large a percentage of total singular values, use a standard svd instead.”
Warning message:
“Keys should be one or more alphanumeric characters followed by an underscore, setting key from pcabrown to pcabrown_”
pcabrown_ 1
Positive: GSTM1, CD8B2, CD70, SMPD3, LRRC2, CPM, GTSF1, PASK, KIR3DL2, CCDC184
HS3ST3B1, DMKN, COL5A1, HIST1H3C, HEATR9, C5, CKLF, C1orf21, EMP2, FGFBP2
AMOT, CCL23, SIGLEC10, NET1, SWAP70, FAM129B, SGO1, NR2F2, TRPV1, EFNB1
Negative: MT1E, MT2A, MT1X, MT1F, CREM, FXYD2, KPNA2, SH2D1A, S100A6, RPS27L
TSC22D1, LGALS3, PDCL3, PRR5L, WDR74, CPNE7, GYG1, HLA-DQA1, TUBB3, FAAH2
CCNA2, FAM83D, TIAM1, RASL11A, EOMES, CXCR5, TYMS, LIMS1, NREP, RHOD
pcabrown_ 2
Positive: CD8B2, GSTM1, CD70, COL1A2, CKLF, COL1A1, LGALS3, TAGLN, GTSF1, IGFBP2
APOBEC3H, RPS27L, HS3ST3B1, IGFBP7, HLA-DQA1, TSC22D1, S100A6, PLPP2, TIAM1, SMPD3
PHLDB2, PASK, EXOC5, LIMS1, CPM, PSMD10, CDKL2, ERAP2, EPYC, TSHR
Negative: CREM, MT1F, MT1E, EOMES, CPNE7, RASL11A, KIR3DL2, TOP2A, C1orf21, CCDC184
FXYD2, KPNA2, WDR74, SH2D1A, CXCR5, RNASE6, SIGLEC10, CCNA2, NREP, FAM83D
C5, SGO1, AGER, FAAH2, RHOD, MT1X, NET1, SWAP70, AMOT, TUBB3
pcabrown_ 3
Positive: LIMS1, EOMES, KPNA2, PRR5L, GYG1, FAAH2, POU3F1, CREM, SH2D1A, WDR74
RASL11A, TSC22D1, IGFBP2, C1orf21, CCDC184, TIAM1, KIR3DL2, ZNF570, FXYD2, LRRC2
PLPP2, ERAP2, NET1, CXCR5, RPS27L, CCNA2, SWAP70, HIST1H4H, HLA-DQA1, TYMS
Negative: MT2A, MT1X, LGALS3, S100A6, MT1E, CKLF, MT1F, COL1A1, CD70, COL1A2
RHOD, EFNB1, COL5A1, TAGLN, GTSF1, CPM, FAM83D, CD8B2, CCL23, FAM129B
C5, EXOC5, DMKN, TRPV1, NREP, AGER, NR2F2, PALLD, SGO1, COL6A1
pcabrown_ 4
Positive: PDCL3, RPS27L, FAM83D, APOBEC3H, CREM, KIR3DL2, PELO, GTSF1, S100A6, CD70
TYMS, HLA-DQA1, EOMES, LGALS3, GYG1, GSTM1, HS3ST3B1, ZNF570, KPNA2, SMPD3
ERAP2, C1orf21, NET1, PGF, HIST1H4H, PSMD10, CPNE7, CCNA2, WDR74, CCDC184
Negative: ANKS1B, FAAH2, COL1A2, TIAM1, CD8B2, LIMS1, IGFBP7, POU3F1, PASK, FXYD2
COL1A1, TUBB3, TAGLN, MT1X, CXCR5, PRR5L, MT1E, TSC22D1, TOP2A, SH2D1A
CPM, PLPP2, TSHR, EFNB1, EMP2, MT1F, RHOD, RNASE6, PHLDB2, TESMIN
pcabrown_ 5
Positive: PELO, LRRC2, MT1X, TIAM1, TUBB3, CPNE7, GYG1, PDCL3, RHOD, ANKS1B
POU3F1, HS3ST3B1, KIR3DL2, WDR74, RPS27L, ERAP2, MT2A, MT1E, NET1, SMPD3
PASK, CD70, APOBEC3H, GSTM1, EOMES, CD8B2, LGALS3, PLPP2, TSHR, FAM83D
Negative: HLA-DQA1, FXYD2, S100A6, SH2D1A, TYMS, CKLF, C1orf21, TSC22D1, EXOC5, PSMD10
TOP2A, KPNA2, CREM, PHLDB2, TAGLN, SWAP70, NREP, TESMIN, EPYC, IGFBP2
ZNF570, IGFBP7, COL1A1, GTSF1, CCDC184, PRR5L, HIST1H3C, AGER, CCNA2, SIGLEC10
Transposing data matrix
Initializing state using k-means centroids initialization
Harmony 1/10
Harmony converged after 1 iterations
Warning message:
“Key ‘harmony_’ taken, using ‘yrtnb_’ instead”
[1] "yellow"
Centering and scaling data matrix
Warning message in irlba(A = t(x = object), nv = npcs, ...):
“You're computing too large a percentage of total singular values, use a standard svd instead.”
Warning message:
“Keys should be one or more alphanumeric characters followed by an underscore, setting key from pcayellow to pcayellow_”
pcayellow_ 1
Positive: RGMA, FCRL6, EGFL6, SLC22A17, HCK, MT1H, RNF130, HIST1H3H, C7orf25, IL13
LRP1, PKP4, SLC25A13, KLHL13, BTBD8, SELENOP, VAV2, ING4, PRSS21, CENPE
RAD54L2, AEN, HLF, FCGR3A, SESTD1, MXI1, CHD7, MAP3K4, NAA16, EEF2K
Negative: ADAM19, CD4, CD52, CD226, CTSB, CD5, CDK6, MGAT5, CCR4, TNFRSF25
CLU, SH3TC1, CLIC1, NETO2, FAS, MBNL2, FRMD4B, LEF1, FKBP5, HIPK2
ANKRD17, PLEKHA5, HAPLN3, CENPF, APOE, UBXN11, CD40LG, DSTYK, HECTD4, ATP10D
pcayellow_ 2
Positive: MBP, CD5, SH3TC1, LRIG1, CD4, CCR4, FCRL6, NAA16, FRMD4B, AKTIP
MXI1, RGMA, AEN, ANKRD17, LEF1, CCDC6, UGGT1, NETO2, MAP3K4, RNF130
POLR1A, SP3, HIST1H3H, FAS, SESTD1, BTBD8, ANKRD26, CCR2, SESN1, EEF2K
Negative: CD52, CLIC1, CD226, ATP10D, TNFRSF25, ADAM19, NSMCE1, MBNL2, CDK6, UBXN11
SEC14L1, HMGB3, FKBP5, NAPSA, HAPLN3, IL9R, BCAR3, FCGR3A, ING4, SLC25A13
HIPK2, VAV2, CTSB, RAD54L2, SYNE3, HECTD4, PRSS21, MGAT5, C7orf25, AGPAT4
pcayellow_ 3
Positive: ATP10D, CENPF, ADAM19, IL9R, NAA16, HMGB3, MGAT5, MXI1, RAD54L2, ANKRD26
FKBP5, RGMA, HIST1H3H, SESN1, EEF2K, TNFRSF25, SEC14L1, CD226, SH3TC1, BCAR3
SP3, MBP, AEN, ING4, LEF1, VAV2, UBXN11, MBNL2, FCGR3A, CD5
Negative: CLIC1, CTSB, UGGT1, CCDC6, CCR4, HIPK2, SESTD1, HAPLN3, RNF130, DSTYK
CD52, APOE, CDK6, CD4, FAS, SYNE3, NETO2, CLU, AKTIP, AGPAT4
NAPSA, FCRL6, CHD7, POLR1A, LRIG1, SELENOP, C7orf25, CD40LG, CCR2, IL13
pcayellow_ 4
Positive: FAS, CCDC6, AEN, SYNE3, SEC14L1, HAPLN3, EEF2K, RAD54L2, ING4, IL9R
HIST1H3H, ANKRD17, CDK6, FRMD4B, CTSB, NAA16, SP3, SESTD1, AGPAT4, MBNL2
CCR2, CENPF, CLIC1, SESN1, DSTYK, ADAM19, AKTIP, LRIG1, CD52, C7orf25
Negative: PLEKHA5, NETO2, CLU, RGMA, APOE, HIPK2, BCAR3, MAP3K4, CHD7, HMGB3
RNF130, LEF1, NAPSA, ANKRD26, FKBP5, UGGT1, HECTD4, CD4, NSMCE1, PRSS21
UBXN11, CD226, MGAT5, CCR4, ATP10D, SELENOP, VAV2, CENPE, LRP1, POLR1A
pcayellow_ 5
Positive: MGAT5, HAPLN3, FCRL6, MAP3K4, CENPE, TNFRSF25, CD4, LEF1, CDK6, MBP
NAA16, FKBP5, RAD54L2, CLU, HIPK2, NETO2, SESTD1, FAS, NAPSA, CCR2
HECTD4, SEC14L1, CD226, PKP4, CD40LG, BCAR3, SESN1, AGPAT4, C7orf25, ADAM19
Negative: ANKRD26, RGMA, APOE, UBXN11, SYNE3, HIST1H3H, LRIG1, SP3, CCR4, ANKRD17
UGGT1, CD52, CCDC6, HMGB3, MXI1, MBNL2, ATP10D, RNF130, CD5, NSMCE1
IL9R, CLIC1, AKTIP, ING4, FCGR3A, FRMD4B, BTBD8, CTSB, CENPF, HCK
Transposing data matrix
Initializing state using k-means centroids initialization
Harmony 1/10
Harmony converged after 1 iterations
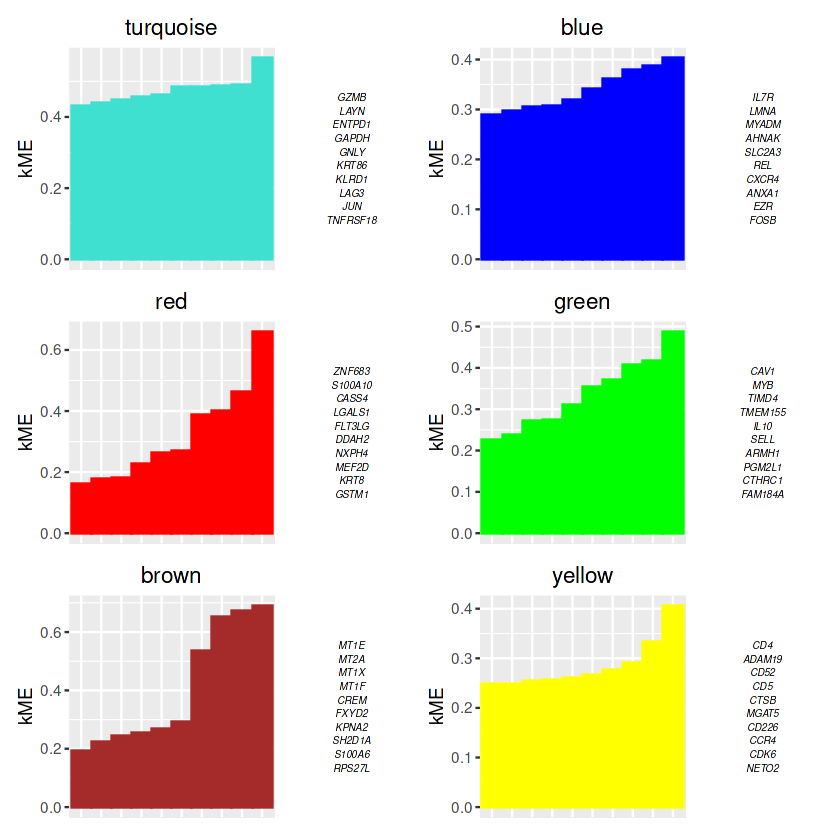
## 画图
p <- PlotKMEs(seurat_obj, ncol=2)
p
# get the module assignment table:
modules <- GetModules(seurat_obj) %>% subset(module != 'grey')
# show the first 6 columns:
head(modules[,1:6])
## 保存模块基因
write.csv(modules,file = "./data/Tex_CD8_modules_wgcna.csv")
| gene_name | module | color | kME_turquoise | kME_blue | kME_grey | |
|---|---|---|---|---|---|---|
| <chr> | <fct> | <chr> | <dbl> | <dbl> | <dbl> | |
| GNLY | GNLY | turquoise | turquoise | 0.48543612 | -0.14525591 | 0.371601226 |
| HBB | HBB | blue | blue | -0.04097688 | 0.07312160 | 0.023877504 |
| CCL4L2 | CCL4L2 | turquoise | turquoise | 0.19331909 | -0.16828326 | -0.003766411 |
| CCL4 | CCL4 | turquoise | turquoise | 0.05447294 | -0.12399817 | -0.078761987 |
| HSPA6 | HSPA6 | turquoise | turquoise | 0.15178861 | 0.00284496 | 0.105700172 |
| XCL1 | XCL1 | blue | blue | 0.12402731 | 0.10158451 | 0.182427043 |
# 枢纽基因
hub_df <- GetHubGenes(seurat_obj, n_hubs = 10)
head(hub_df)
write.csv(hub_df,file = "./data/Tex_CD8_hub_wgcna.csv")
## 保存分析的结果
saveRDS(seurat_obj, file='./data/Tex_CD8_hdWGCNA_object.rds')
## 绘制枢纽基因网络图
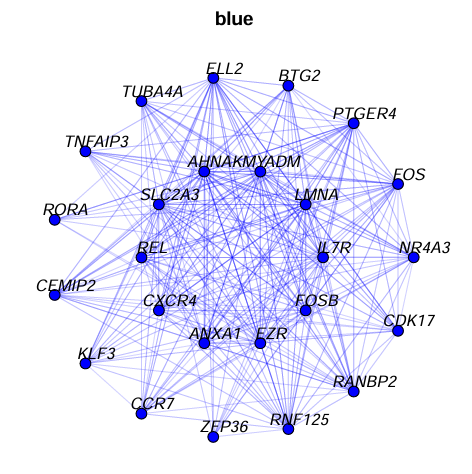
ModuleNetworkPlot(
seurat_obj,
outdir = "modulenetworks"
)
ModuleNetworkPlot(
seurat_obj,
outdir = "modulenetworks2",
n_inner = 20,
n_outer = 30,
n_conns = Inf,
plot_size = c(10,10),
vertex.label.cex = 1
)
## 这样两个文件夹内就有了模块的图片
| gene_name | module | kME | |
|---|---|---|---|
| <chr> | <fct> | <dbl> | |
| 1 | GZMB | turquoise | 0.5663421 |
| 2 | LAYN | turquoise | 0.4908385 |
| 3 | ENTPD1 | turquoise | 0.4876040 |
| 4 | GAPDH | turquoise | 0.4856226 |
| 5 | GNLY | turquoise | 0.4854361 |
| 6 | KRT86 | turquoise | 0.4630110 |
Writing output files to modulenetworks[1] "turquoise"
[1] "blue"
[1] "red"
[1] "green"
[1] "brown"
[1] "yellow"
Writing output files to modulenetworks2[1] "turquoise"
[1] "blue"
[1] "red"
[1] "green"
[1] "brown"
[1] "yellow"
hub网络图如下