Seurat单细胞处理流程之八:pySCENIC转录因子分析-python可视化
# import dependencies
import loompy as lp
import json
import base64
import zlib
import pandas as pd
import scanpy as sc
from pyscenic.plotting import plot_binarization
from pyscenic.export import add_scenic_metadata
import seaborn as sns
import matplotlib as mpl
import matplotlib.pyplot as plt
from pyscenic.rss import regulon_specificity_scores
from pyscenic.plotting import plot_rss
import matplotlib.pyplot as plt
from adjustText import adjust_text
import seaborn as sns
from pyscenic.binarization import binarize
OMP: Info #276: omp_set_nested routine deprecated, please use omp_set_max_active_levels instead.
sc.settings.verbosity = 3 # verbosity: errors (0), warnings (1), info (2), hints (3)
sc.logging.print_versions()
sc.set_figure_params(dpi=300, fontsize=10, dpi_save=600)
-----
anndata 0.8.0
scanpy 1.9.3
-----
PIL 9.5.0
adjustText 1.3.0
attr 22.1.0
backcall 0.2.0
beta_ufunc NA
binom_ufunc NA
boltons NA
cffi 1.15.1
cloudpickle 2.2.1
ctxcore 0.2.0
cycler 0.10.0
cython_runtime NA
cytoolz 0.12.3
dask 2022.02.0
dateutil 2.8.2
debugpy 1.5.1
decorator 5.1.1
defusedxml 0.7.1
entrypoints 0.4
frozendict 2.4.5
fsspec 2023.1.0
h5py 3.8.0
ipykernel 6.15.2
ipython_genutils 0.2.0
ipywidgets 7.6.5
jedi 0.18.1
jinja2 3.1.2
joblib 1.3.2
jupyter_server 1.23.4
kiwisolver 1.4.5
llvmlite 0.39.1
loompy 3.0.7
markupsafe 2.1.1
matplotlib 3.5.3
matplotlib_inline 0.1.6
mkl 2.4.0
mpl_toolkits NA
natsort 8.4.0
nbinom_ufunc NA
networkx 2.6.3
numba 0.56.4
numexpr 2.8.6
numpy 1.21.5
numpy_groupies 0.9.22
packaging 22.0
pandas 1.3.5
parso 0.8.3
pexpect 4.8.0
pickleshare 0.7.5
pkg_resources NA
prompt_toolkit 3.0.36
psutil 5.9.0
ptyprocess 0.7.0
pyarrow 12.0.1
pycparser 2.21
pydev_ipython NA
pydevconsole NA
pydevd 2.6.0
pydevd_concurrency_analyser NA
pydevd_file_utils NA
pydevd_plugins NA
pydevd_tracing NA
pygments 2.11.2
pyparsing 3.1.4
pyscenic 0.12.1
pytz 2022.7
scipy 1.7.3
seaborn 0.12.2
session_info 1.0.0
six 1.16.0
sklearn 1.0.2
statsmodels 0.13.5
storemagic NA
tblib 2.0.0
threadpoolctl 3.1.0
tlz 0.12.3
toolz 0.12.1
tornado 6.2
tqdm 4.66.5
traitlets 5.7.1
typing_extensions NA
vscode NA
wcwidth 0.2.5
yaml 6.0.1
zipp NA
zmq 23.2.0
-----
IPython 7.31.1
jupyter_client 7.4.9
jupyter_core 4.11.2
jupyterlab 3.5.3
notebook 6.5.2
-----
Python 3.7.16 (default, Jan 17 2023, 22:20:44) [GCC 11.2.0]
Linux-5.15.0-130-generic-x86_64-with-debian-bullseye-sid
-----
Session information updated at 2025-04-03 14:20
## 读取pyscenic计算之后的loom文件
sample_SCENIC = 'out_pbmc_SCENIC.loom'
# 提取转录因子AUC评分,这里得到的文件和我们在R里面第一部分提取的文件一样
lf = lp.connect(sample_SCENIC, mode='r+', validate=False )
auc_mtx = pd.DataFrame(lf.ca.RegulonsAUC, index=lf.ca.CellID)
lf.close()
auc_mtx
| AHR(+) | APEX1(+) | ARID3A(+) | ARNTL(+) | ATF1(+) | ATF3(+) | ATF4(+) | ATF6(+) | ATF6B(+) | BACH1(+) | ... | ZNF583(+) | ZNF585A(+) | ZNF689(+) | ZNF714(+) | ZNF76(+) | ZNF785(+) | ZNF814(+) | ZNF880(+) | ZNF92(+) | ZXDB(+) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AAACATACAACCAC-1 | 0.000000 | 0.145773 | 0.010933 | 0.024619 | 0.000000 | 0.045329 | 0.033024 | 0.000000 | 0.000000 | 0.037362 | ... | 0.036576 | 0.000000 | 0.057726 | 0.000000 | 0.139942 | 0.018278 | 0.024690 | 0.080661 | 0.060841 | 0.000000 |
| AAACATTGAGCTAC-1 | 0.000000 | 0.232264 | 0.000000 | 0.005251 | 0.000000 | 0.059722 | 0.052328 | 0.000000 | 0.000000 | 0.024414 | ... | 0.095150 | 0.013767 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.015579 | 0.000000 | 0.047414 | 0.012222 |
| AAACATTGATCAGC-1 | 0.032070 | 0.072886 | 0.002936 | 0.013578 | 0.048001 | 0.050850 | 0.047495 | 0.000000 | 0.000000 | 0.014038 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.049675 | 0.048014 | 0.000000 | 0.005677 | 0.000000 |
| AAACCGTGCTTCCG-1 | 0.040233 | 0.187075 | 0.096007 | 0.002996 | 0.000000 | 0.126092 | 0.072065 | 0.000000 | 0.000000 | 0.028959 | ... | 0.000000 | 0.054260 | 0.087464 | 0.042760 | 0.000000 | 0.012895 | 0.035168 | 0.000000 | 0.077797 | 0.000000 |
| AAACCGTGTATGCG-1 | 0.000000 | 0.135083 | 0.006317 | 0.016629 | 0.025718 | 0.048888 | 0.030091 | 0.018542 | 0.057945 | 0.043377 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.036534 | 0.000000 | 0.000000 | 0.000000 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| TTTCGAACTCTCAT-1 | 0.000000 | 0.140428 | 0.036787 | 0.013187 | 0.000000 | 0.132603 | 0.085193 | 0.000000 | 0.000000 | 0.029779 | ... | 0.010602 | 0.000000 | 0.061516 | 0.103499 | 0.000000 | 0.044629 | 0.029883 | 0.000000 | 0.082707 | 0.017941 |
| TTTCTACTGAGGCA-1 | 0.024198 | 0.149417 | 0.017290 | 0.015347 | 0.020512 | 0.047221 | 0.056524 | 0.000000 | 0.000000 | 0.006897 | ... | 0.041744 | 0.000000 | 0.105831 | 0.000000 | 0.000000 | 0.000000 | 0.096939 | 0.000000 | 0.038284 | 0.042835 |
| TTTCTACTTCCTCG-1 | 0.081050 | 0.115403 | 0.007309 | 0.011081 | 0.000000 | 0.064134 | 0.045110 | 0.001050 | 0.000000 | 0.017235 | ... | 0.138616 | 0.000000 | 0.195044 | 0.000000 | 0.000000 | 0.084212 | 0.051567 | 0.050373 | 0.091300 | 0.000000 |
| TTTGCATGAGAGGC-1 | 0.000000 | 0.000000 | 0.002045 | 0.016075 | 0.000000 | 0.046311 | 0.033722 | 0.011020 | 0.007653 | 0.015398 | ... | 0.116088 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.050684 | 0.045554 | 0.000000 | 0.098588 | 0.000000 |
| TTTGCATGCCTCAC-1 | 0.000000 | 0.144801 | 0.001559 | 0.012526 | 0.000000 | 0.033144 | 0.042407 | 0.000175 | 0.000000 | 0.029363 | ... | 0.019746 | 0.022514 | 0.000000 | 0.049320 | 0.000000 | 0.076362 | 0.080357 | 0.000000 | 0.026086 | 0.025903 |
2638 rows × 272 columns
#Calculate RSS,用于后续的可视化,这里是按照细胞类型
pbmc = sc.read_h5ad("../../scanpy_单细胞流程/pbmc_test/adata_注释后.h5ad")
rss_cellType = regulon_specificity_scores( auc_mtx, pbmc.obs.celltype )
rss_cellType
| AHR(+) | APEX1(+) | ARID3A(+) | ARNTL(+) | ATF1(+) | ATF3(+) | ATF4(+) | ATF6(+) | ATF6B(+) | BACH1(+) | ... | ZNF583(+) | ZNF585A(+) | ZNF689(+) | ZNF714(+) | ZNF76(+) | ZNF785(+) | ZNF814(+) | ZNF880(+) | ZNF92(+) | ZXDB(+) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD4 T | 0.371006 | 0.494335 | 0.362952 | 0.514898 | 0.320398 | 0.404762 | 0.452648 | 0.331502 | 0.316784 | 0.391548 | ... | 0.328759 | 0.234516 | 0.289599 | 0.339651 | 0.235871 | 0.424768 | 0.417361 | 0.319822 | 0.411642 | 0.387903 |
| B | 0.260599 | 0.297163 | 0.259716 | 0.279114 | 0.239077 | 0.271071 | 0.277191 | 0.256355 | 0.269571 | 0.276397 | ... | 0.353579 | 0.273430 | 0.204549 | 0.261727 | 0.190374 | 0.282572 | 0.280896 | 0.287629 | 0.369873 | 0.247092 |
| FCGR3A+ Monocytes | 0.219350 | 0.240290 | 0.274068 | 0.212988 | 0.224245 | 0.284827 | 0.262481 | 0.194940 | 0.209400 | 0.270493 | ... | 0.226854 | 0.191498 | 0.204842 | 0.221876 | 0.173565 | 0.210461 | 0.259366 | 0.186186 | 0.231726 | 0.197176 |
| NK | 0.230367 | 0.222013 | 0.207571 | 0.254317 | 0.221009 | 0.226006 | 0.222470 | 0.213226 | 0.262290 | 0.219938 | ... | 0.245548 | 0.205701 | 0.202699 | 0.208693 | 0.242466 | 0.235943 | 0.234619 | 0.202609 | 0.208136 | 0.196424 |
| CD8 T | 0.249879 | 0.268242 | 0.238305 | 0.289165 | 0.249358 | 0.255727 | 0.263332 | 0.235437 | 0.237345 | 0.246176 | ... | 0.266994 | 0.206734 | 0.218780 | 0.228390 | 0.331778 | 0.276096 | 0.260976 | 0.241157 | 0.306110 | 0.219693 |
| CD14+ Monocytes | 0.289323 | 0.317279 | 0.455322 | 0.295596 | 0.245026 | 0.448463 | 0.406507 | 0.270345 | 0.270238 | 0.452946 | ... | 0.247070 | 0.224899 | 0.261631 | 0.274461 | 0.187960 | 0.260316 | 0.295780 | 0.231593 | 0.303824 | 0.277495 |
| Dendritic | 0.174583 | 0.192224 | 0.201737 | 0.182372 | 0.177555 | 0.201129 | 0.197314 | 0.172322 | 0.187669 | 0.201658 | ... | 0.183138 | 0.173830 | 0.171719 | 0.191358 | 0.167445 | 0.183616 | 0.186164 | 0.190682 | 0.198571 | 0.179925 |
| Megakaryocytes | 0.180727 | 0.173432 | 0.183540 | 0.177623 | 0.169538 | 0.176384 | 0.179265 | 0.193616 | 0.180323 | 0.176580 | ... | 0.169004 | 0.167445 | 0.176624 | 0.171554 | 0.172148 | 0.170093 | 0.176444 | 0.167445 | 0.170680 | 0.169616 |
8 rows × 272 columns
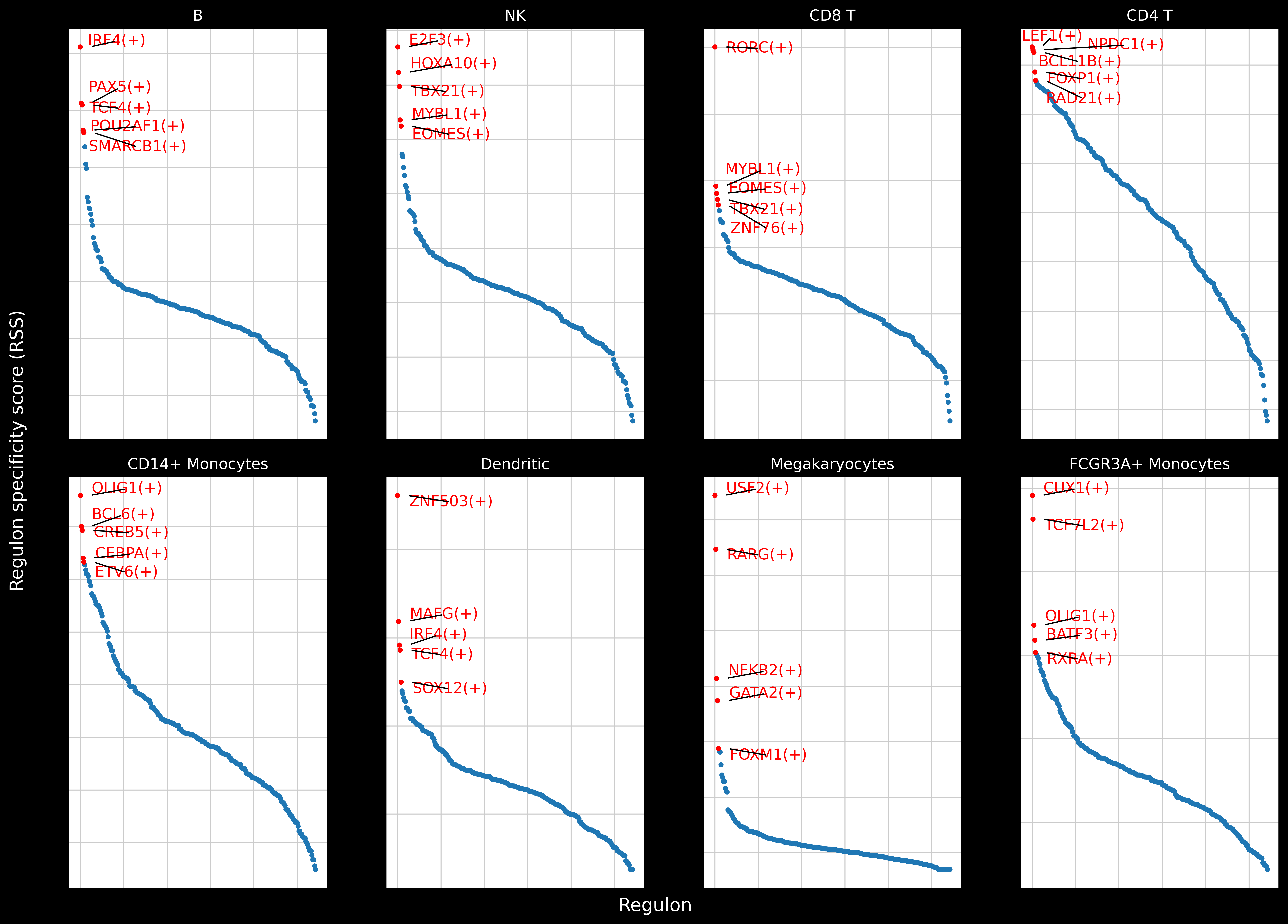
import matplotlib.pyplot as plt
from adjustText import adjust_text
celltype = ["B","NK","CD8 T","CD4 T",
"CD14+ Monocytes","Dendritic","Megakaryocytes","FCGR3A+ Monocytes"]
fig = plt.figure(figsize=(14, 10)) # 图的高低
for c, num in zip(celltype, range(1, len(celltype) + 1)):
x = rss_cellType.T[c]
ax = fig.add_subplot(2, 4, num) # 图的排列
# 动态设置 top_n,防止超过实际数量
plot_rss(rss_cellType, c, top_n=min(5, len(rss_cellType.T[c])), max_n=None, ax=ax)
ax.set_ylim(x.min() - (x.max() - x.min()) * 0.05, x.max() + (x.max() - x.min()) * 0.05)
# 调整字体大小
for t in ax.texts:
t.set_fontsize(12)
ax.set_ylabel('')
ax.set_xlabel('')
# 筛选出包含正号 '(+)' 的文本,避免调整非标注文本
texts_to_adjust = [t for t in ax.texts if '(+)' in t.get_text()]
if texts_to_adjust: # 确保有文本才调整
adjust_text(texts_to_adjust, autoalign='xy', ha='right', va='bottom',
arrowprops=dict(arrowstyle='-', color='black'), precision=0.001)
fig.text(0.5, 0.0, 'Regulon', ha='center', va='center', size='x-large')
fig.text(0.00, 0.5, 'Regulon specificity score (RSS)', ha='center', va='center', rotation='vertical', size='x-large')
plt.tight_layout()
plt.rcParams.update({
'figure.autolayout': True,
'figure.titlesize': 'large',
'axes.labelsize': 'medium',
'axes.titlesize': 'large',
'xtick.labelsize': 'medium',
'ytick.labelsize': 'medium'
})
plt.savefig("rank_RSS_top5.pdf", dpi=600, bbox_inches="tight")
plt.show()
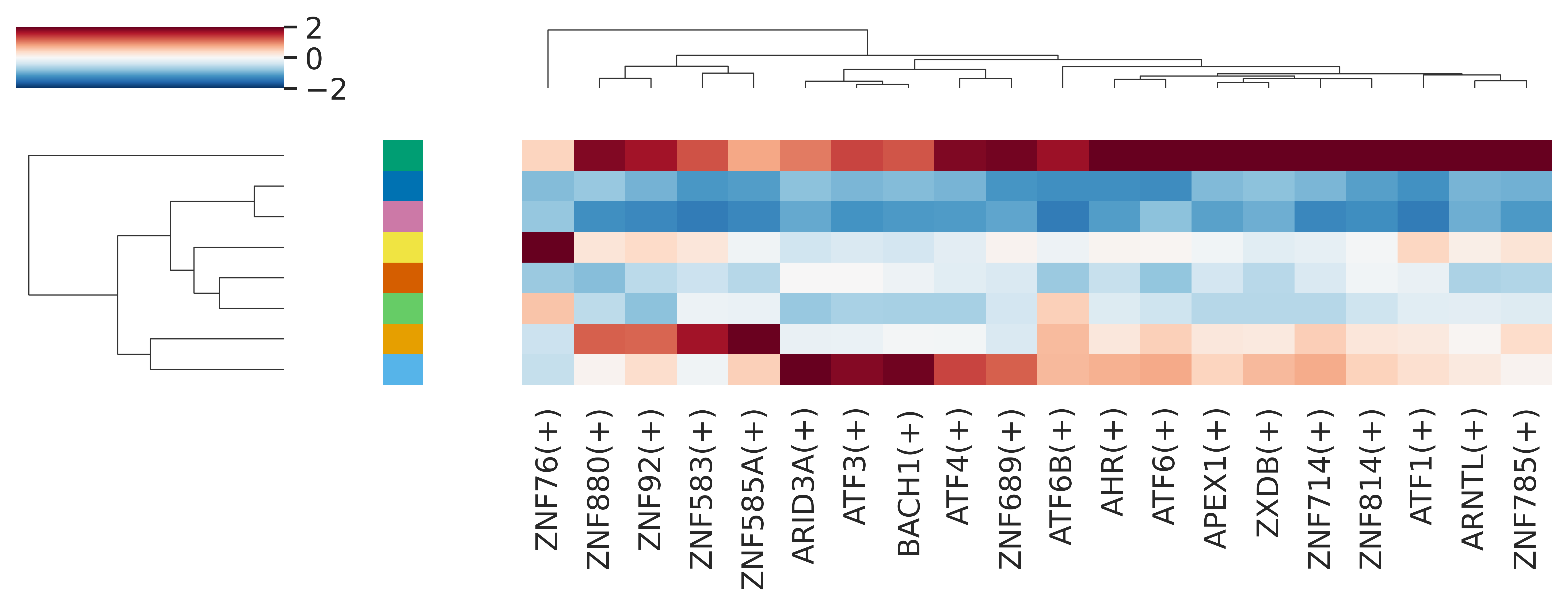
## tf热图
#对于RSS矩阵,我们也可以像R中的可视化一样,我们做气泡图或者热图
#计算下Z-score
rss_cellType_Z = pd.DataFrame( index=rss_cellType.index )
for col in list(rss_cellType.columns):
rss_cellType_Z[col] = ( rss_cellType[col] - rss_cellType[col].mean()) / rss_cellType[col].std(ddof=0)
rss_cellType_Z.sort_index(inplace=True)
rss_cellType_Z
/home/zhaozm/anaconda3/envs/pyscenic/lib/python3.7/site-packages/ipykernel_launcher.py:6: PerformanceWarning: DataFrame is highly fragmented. This is usually the result of calling `frame.insert` many times, which has poor performance. Consider joining all columns at once using pd.concat(axis=1) instead. To get a de-fragmented frame, use `newframe = frame.copy()`
| AHR(+) | APEX1(+) | ARID3A(+) | ARNTL(+) | ATF1(+) | ATF3(+) | ATF4(+) | ATF6(+) | ATF6B(+) | BACH1(+) | ... | ZNF583(+) | ZNF585A(+) | ZNF689(+) | ZNF714(+) | ZNF76(+) | ZNF785(+) | ZNF814(+) | ZNF880(+) | ZNF92(+) | ZXDB(+) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | 0.230048 | 0.227674 | -0.152153 | 0.033440 | 0.189383 | -0.139034 | -0.059820 | 0.473134 | 0.635161 | -0.034311 | ... | 1.683834 | 1.972466 | -0.310758 | 0.489999 | -0.426368 | 0.362381 | 0.241358 | 1.196053 | 1.169846 | 0.189518 |
| CD14+ Monocytes | 0.715243 | 0.440301 | 2.105071 | 0.197723 | 0.325112 | 1.837960 | 1.357026 | 0.762349 | 0.650349 | 1.938962 | ... | -0.092537 | 0.469083 | 1.198087 | 0.744527 | -0.472468 | 0.064655 | 0.453361 | 0.064656 | 0.354817 | 0.644069 |
| CD4 T | 2.094955 | 2.311712 | 1.039159 | 2.383599 | 2.044672 | 1.350924 | 1.862568 | 2.026639 | 1.711183 | 1.252724 | ... | 1.269887 | 0.766988 | 1.937369 | 2.047613 | 0.442470 | 2.264576 | 2.185080 | 1.846057 | 1.685269 | 2.294761 |
| CD8 T | 0.048988 | -0.078006 | -0.399233 | 0.133622 | 0.423954 | -0.310035 | -0.211668 | 0.040717 | -0.099315 | -0.372090 | ... | 0.239752 | -0.093630 | 0.065414 | -0.176372 | 2.273967 | 0.275742 | -0.042372 | 0.257766 | 0.383023 | -0.220109 |
| Dendritic | -1.222861 | -0.881488 | -0.821212 | -0.930829 | -1.214203 | -0.918519 | -0.934991 | -1.264052 | -1.231476 | -0.869669 | ... | -1.158793 | -1.112942 | -1.178548 | -0.916601 | -0.864224 | -0.961383 | -1.107943 | -0.761347 | -0.943982 | -0.814669 |
| FCGR3A+ Monocytes | -0.466699 | -0.373446 | 0.013456 | -0.625669 | -0.149002 | 0.014275 | -0.220995 | -0.796479 | -0.736208 | -0.100307 | ... | -0.429706 | -0.565603 | -0.303010 | -0.306586 | -0.747367 | -0.602279 | -0.065309 | -0.852139 | -0.534859 | -0.556764 |
| Megakaryocytes | -1.119079 | -1.080113 | -1.031198 | -0.978165 | -1.397095 | -1.194294 | -1.132750 | -0.823853 | -1.398899 | -1.149957 | ... | -1.394522 | -1.310713 | -1.048903 | -1.312469 | -0.774428 | -1.142295 | -1.246392 | -1.230512 | -1.288157 | -0.968806 |
| NK | -0.280595 | -0.566634 | -0.753889 | -0.213721 | -0.222820 | -0.641276 | -0.659369 | -0.418454 | 0.469205 | -0.665352 | ... | -0.117915 | -0.125650 | -0.359651 | -0.570111 | 0.568417 | -0.261397 | -0.417783 | -0.520535 | -0.825955 | -0.568001 |
8 rows × 272 columns
print(rss_cellType_Z.columns)
print(rss_cellType_Z.shape) # (行数, 列数)
print(len(colors)) # 应该等于 8
Index(['AHR(+)', 'APEX1(+)', 'ARID3A(+)', 'ARNTL(+)', 'ATF1(+)', 'ATF3(+)',
'ATF4(+)', 'ATF6(+)', 'ATF6B(+)', 'BACH1(+)',
...
'ZNF583(+)', 'ZNF585A(+)', 'ZNF689(+)', 'ZNF714(+)', 'ZNF76(+)',
'ZNF785(+)', 'ZNF814(+)', 'ZNF880(+)', 'ZNF92(+)', 'ZXDB(+)'],
dtype='object', length=272)
(8, 272)
7
colors = ["#E69F00","#56B4E9","#009E73","#F0E442","#0072B2","#D55E00","#CC79A7",'#66CC66']#行注释颜色
sns.set(font_scale=1.2)
TF_plot = ['AHR(+)', 'APEX1(+)', 'ARID3A(+)', 'ARNTL(+)', 'ATF1(+)', 'ATF3(+)',
'ATF4(+)', 'ATF6(+)', 'ATF6B(+)', 'BACH1(+)','ZNF583(+)', 'ZNF585A(+)', 'ZNF689(+)', 'ZNF714(+)', 'ZNF76(+)',
'ZNF785(+)', 'ZNF814(+)', 'ZNF880(+)', 'ZNF92(+)', 'ZXDB(+)']#选择需要呈现的TF
g = sns.clustermap(rss_cellType_Z[TF_plot],
annot=False, square=False,
linecolor='black',
yticklabels=False, xticklabels=True,
vmin=-2, vmax=2, #颜色范围
row_colors=colors,cmap="RdBu_r", figsize=(10,4) )#颜色,图尺寸
g.cax.set_visible(True)#显示legend
g.ax_heatmap.set_ylabel('')
g.ax_heatmap.set_xlabel('')
Text(0.5, -239.86666666666676, '')
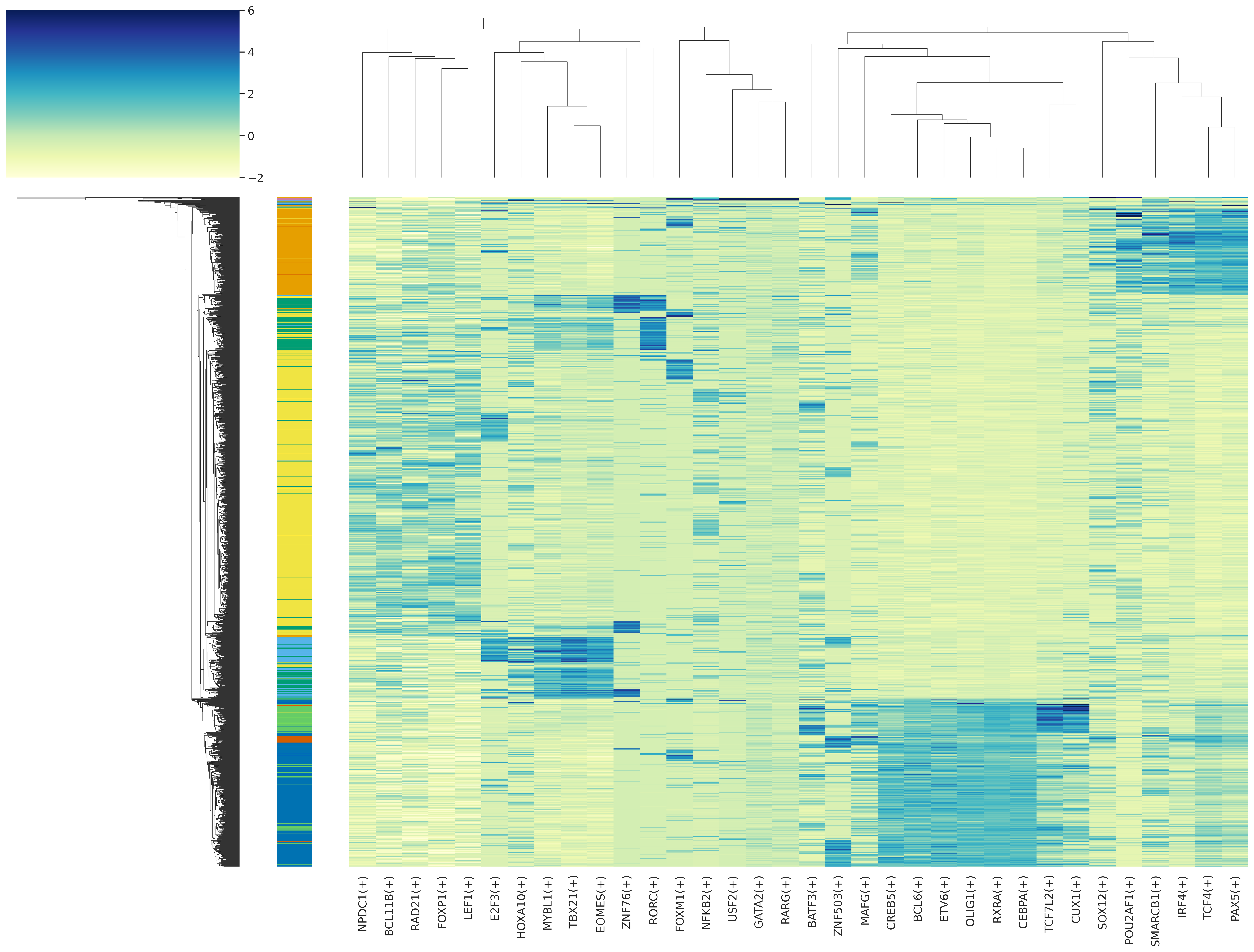
#做所有细胞的热图
#挑选每个细胞类型中top5的TF,当然了,我们也可以只选择自己感兴趣的TF
topreg = []
for i,c in enumerate(celltype):
topreg.extend(
list(rss_cellType.T[c].sort_values(ascending=False)[:5].index)
)
topreg = list(set(topreg))
auc_mtx_Z = pd.DataFrame( index=auc_mtx.index )
for col in list(auc_mtx.columns):
auc_mtx_Z[ col ] = ( auc_mtx[col] - auc_mtx[col].mean()) / auc_mtx[col].std(ddof=0)
/home/zhaozm/anaconda3/envs/pyscenic/lib/python3.7/site-packages/ipykernel_launcher.py:3: PerformanceWarning: DataFrame is highly fragmented. This is usually the result of calling `frame.insert` many times, which has poor performance. Consider joining all columns at once using pd.concat(axis=1) instead. To get a de-fragmented frame, use `newframe = frame.copy()`
This is separate from the ipykernel package so we can avoid doing imports until
colors = ["#E69F00","#56B4E9","#009E73","#F0E442","#0072B2","#D55E00","#CC79A7",'#66CC66']
colorsd = dict( zip( celltype, colors ))
colormap = [ colorsd[x] for x in pbmc.obs.celltype]
sns.set(font_scale=1.2)
g = sns.clustermap(auc_mtx_Z[topreg], annot=False, square=False, linecolor='black',
yticklabels=False, xticklabels=True, vmin=-2, vmax=6, row_colors=colormap,
cmap="YlGnBu", figsize=(21,16) )
g.cax.set_visible(True)
g.ax_heatmap.set_ylabel('')
g.ax_heatmap.set_xlabel('')
plt.savefig("TF-heatmap-top5.pdf", dpi=600, bbox_inches = "tight")
/home/zhaozm/anaconda3/envs/pyscenic/lib/python3.7/site-packages/seaborn/matrix.py:560: UserWarning: Clustering large matrix with scipy. Installing `fastcluster` may give better performance.
warnings.warn(msg)
# 可视化3:TF活性聚类图
#将auc添加到单细胞
add_scenic_metadata(pbmc, auc_mtx)
AnnData object with n_obs × n_vars = 2638 × 1838
obs: 'n_genes', 'n_genes_by_counts', 'total_counts', 'total_counts_mt', 'pct_counts_mt', 'leiden', 'celltype', 'Regulon(AHR(+))', 'Regulon(APEX1(+))', 'Regulon(ARID3A(+))', 'Regulon(ARNTL(+))', 'Regulon(ATF1(+))', 'Regulon(ATF3(+))', 'Regulon(ATF4(+))', 'Regulon(ATF6(+))', 'Regulon(ATF6B(+))', 'Regulon(BACH1(+))', 'Regulon(BACH2(+))', 'Regulon(BATF(+))', 'Regulon(BATF3(+))', 'Regulon(BCL11A(+))', 'Regulon(BCL11B(+))', 'Regulon(BCL3(+))', 'Regulon(BCL6(+))', 'Regulon(BCLAF1(+))', 'Regulon(BDP1(+))', 'Regulon(BHLHE40(+))', 'Regulon(BRCA1(+))', 'Regulon(BRF1(+))', 'Regulon(BRF2(+))', 'Regulon(CEBPA(+))', 'Regulon(CEBPB(+))', 'Regulon(CEBPD(+))', 'Regulon(CEBPE(+))', 'Regulon(CEBPG(+))', 'Regulon(CENPB(+))', 'Regulon(CIC(+))', 'Regulon(CLOCK(+))', 'Regulon(CREB3(+))', 'Regulon(CREB3L2(+))', 'Regulon(CREB3L4(+))', 'Regulon(CREB5(+))', 'Regulon(CREBZF(+))', 'Regulon(CUX1(+))', 'Regulon(E2F3(+))', 'Regulon(E2F6(+))', 'Regulon(E4F1(+))', 'Regulon(EBF1(+))', 'Regulon(EGR2(+))', 'Regulon(ELF1(+))', 'Regulon(ELF2(+))', 'Regulon(ELF4(+))', 'Regulon(ELK1(+))', 'Regulon(ELK3(+))', 'Regulon(ELK4(+))', 'Regulon(EOMES(+))', 'Regulon(EP300(+))', 'Regulon(ERF(+))', 'Regulon(ESRRA(+))', 'Regulon(ETS1(+))', 'Regulon(ETS2(+))', 'Regulon(ETV2(+))', 'Regulon(ETV3(+))', 'Regulon(ETV5(+))', 'Regulon(ETV6(+))', 'Regulon(ETV7(+))', 'Regulon(EZH2(+))', 'Regulon(FLI1(+))', 'Regulon(FOS(+))', 'Regulon(FOSB(+))', 'Regulon(FOSL1(+))', 'Regulon(FOSL2(+))', 'Regulon(FOXD2(+))', 'Regulon(FOXJ3(+))', 'Regulon(FOXM1(+))', 'Regulon(FOXN2(+))', 'Regulon(FOXO3(+))', 'Regulon(FOXO4(+))', 'Regulon(FOXP1(+))', 'Regulon(FOXP4(+))', 'Regulon(GABPA(+))', 'Regulon(GABPB1(+))', 'Regulon(GATA2(+))', 'Regulon(GATA3(+))', 'Regulon(GMEB2(+))', 'Regulon(GTF2IRD1(+))', 'Regulon(GTF3C2(+))', 'Regulon(HBP1(+))', 'Regulon(HCFC1(+))', 'Regulon(HINFP(+))', 'Regulon(HMGB1(+))', 'Regulon(HMGN3(+))', 'Regulon(HOXA10(+))', 'Regulon(HOXA9(+))', 'Regulon(HSF2(+))', 'Regulon(HSF4(+))', 'Regulon(IRF1(+))', 'Regulon(IRF3(+))', 'Regulon(IRF4(+))', 'Regulon(IRF5(+))', 'Regulon(IRF7(+))', 'Regulon(IRF8(+))', 'Regulon(IRF9(+))', 'Regulon(JDP2(+))', 'Regulon(JUN(+))', 'Regulon(JUNB(+))', 'Regulon(JUND(+))', 'Regulon(KDM4B(+))', 'Regulon(KDM5B(+))', 'Regulon(KLF10(+))', 'Regulon(KLF2(+))', 'Regulon(KLF3(+))', 'Regulon(KLF4(+))', 'Regulon(KLF6(+))', 'Regulon(LEF1(+))', 'Regulon(MAF(+))', 'Regulon(MAFB(+))', 'Regulon(MAFF(+))', 'Regulon(MAFG(+))', 'Regulon(MAX(+))', 'Regulon(MECP2(+))', 'Regulon(MEF2A(+))', 'Regulon(MEF2D(+))', 'Regulon(MEOX1(+))', 'Regulon(MGA(+))', 'Regulon(MITF(+))', 'Regulon(MLX(+))', 'Regulon(MSC(+))', 'Regulon(MXD3(+))', 'Regulon(MXI1(+))', 'Regulon(MYB(+))', 'Regulon(MYBL1(+))', 'Regulon(MYBL2(+))', 'Regulon(MYC(+))', 'Regulon(MYEF2(+))', 'Regulon(MZF1(+))', 'Regulon(NEUROD2(+))', 'Regulon(NFE2(+))', 'Regulon(NFE2L1(+))', 'Regulon(NFE2L3(+))', 'Regulon(NFIC(+))', 'Regulon(NFIL3(+))', 'Regulon(NFKB2(+))', 'Regulon(NFYA(+))', 'Regulon(NFYB(+))', 'Regulon(NPAS2(+))', 'Regulon(NPDC1(+))', 'Regulon(NR1D1(+))', 'Regulon(NR1H2(+))', 'Regulon(NR1H3(+))', 'Regulon(NR2C1(+))', 'Regulon(NR2C2(+))', 'Regulon(NR2F6(+))', 'Regulon(NR3C1(+))', 'Regulon(NUAK1(+))', 'Regulon(OLIG1(+))', 'Regulon(PAX5(+))', 'Regulon(PAX8(+))', 'Regulon(PBX3(+))', 'Regulon(PHF8(+))', 'Regulon(PKNOX1(+))', 'Regulon(PML(+))', 'Regulon(POLE3(+))', 'Regulon(POLR2A(+))', 'Regulon(POLR3A(+))', 'Regulon(POU2AF1(+))', 'Regulon(POU2F1(+))', 'Regulon(POU6F1(+))', 'Regulon(PPARA(+))', 'Regulon(PPARD(+))', 'Regulon(PSMD12(+))', 'Regulon(RAD21(+))', 'Regulon(RARA(+))', 'Regulon(RARG(+))', 'Regulon(RBBP5(+))', 'Regulon(RCOR1(+))', 'Regulon(RELA(+))', 'Regulon(RELB(+))', 'Regulon(REST(+))', 'Regulon(RFX1(+))', 'Regulon(RFX2(+))', 'Regulon(RFX3(+))', 'Regulon(RFX5(+))', 'Regulon(RFX7(+))', 'Regulon(RFXAP(+))', 'Regulon(RORC(+))', 'Regulon(RREB1(+))', 'Regulon(RUNX1(+))', 'Regulon(RUNX2(+))', 'Regulon(RUNX3(+))', 'Regulon(RXRA(+))', 'Regulon(RXRB(+))', 'Regulon(SAP30(+))', 'Regulon(SETDB1(+))', 'Regulon(SMAD1(+))', 'Regulon(SMARCA4(+))', 'Regulon(SMARCB1(+))', 'Regulon(SNAI1(+))', 'Regulon(SOX12(+))', 'Regulon(SOX13(+))', 'Regulon(SOX7(+))', 'Regulon(SP1(+))', 'Regulon(SP2(+))', 'Regulon(SP4(+))', 'Regulon(SPI1(+))', 'Regulon(SPIB(+))', 'Regulon(SREBF1(+))', 'Regulon(SREBF2(+))', 'Regulon(SRF(+))', 'Regulon(STAT1(+))', 'Regulon(STAT2(+))', 'Regulon(STAT3(+))', 'Regulon(STAT5A(+))', 'Regulon(TAF1(+))', 'Regulon(TAF7(+))', 'Regulon(TAL1(+))', 'Regulon(TBP(+))', 'Regulon(TBX19(+))', 'Regulon(TBX21(+))', 'Regulon(TCF12(+))', 'Regulon(TCF3(+))', 'Regulon(TCF4(+))', 'Regulon(TCF7L2(+))', 'Regulon(TERF2(+))', 'Regulon(TFAP4(+))', 'Regulon(TFEB(+))', 'Regulon(TGIF2(+))', 'Regulon(THAP11(+))', 'Regulon(TP53(+))', 'Regulon(TRIM28(+))', 'Regulon(USF1(+))', 'Regulon(USF2(+))', 'Regulon(VDR(+))', 'Regulon(XBP1(+))', 'Regulon(XRCC4(+))', 'Regulon(YY2(+))', 'Regulon(ZBTB14(+))', 'Regulon(ZBTB3(+))', 'Regulon(ZBTB33(+))', 'Regulon(ZBTB6(+))', 'Regulon(ZBTB7A(+))', 'Regulon(ZEB1(+))', 'Regulon(ZFP1(+))', 'Regulon(ZFX(+))', 'Regulon(ZFY(+))', 'Regulon(ZMIZ1(+))', 'Regulon(ZNF121(+))', 'Regulon(ZNF133(+))', 'Regulon(ZNF143(+))', 'Regulon(ZNF169(+))', 'Regulon(ZNF200(+))', 'Regulon(ZNF224(+))', 'Regulon(ZNF234(+))', 'Regulon(ZNF248(+))', 'Regulon(ZNF274(+))', 'Regulon(ZNF282(+))', 'Regulon(ZNF286A(+))', 'Regulon(ZNF317(+))', 'Regulon(ZNF329(+))', 'Regulon(ZNF337(+))', 'Regulon(ZNF383(+))', 'Regulon(ZNF43(+))', 'Regulon(ZNF442(+))', 'Regulon(ZNF496(+))', 'Regulon(ZNF503(+))', 'Regulon(ZNF549(+))', 'Regulon(ZNF567(+))', 'Regulon(ZNF568(+))', 'Regulon(ZNF569(+))', 'Regulon(ZNF583(+))', 'Regulon(ZNF585A(+))', 'Regulon(ZNF689(+))', 'Regulon(ZNF714(+))', 'Regulon(ZNF76(+))', 'Regulon(ZNF785(+))', 'Regulon(ZNF814(+))', 'Regulon(ZNF880(+))', 'Regulon(ZNF92(+))', 'Regulon(ZXDB(+))'
var: 'gene_ids', 'n_cells', 'mt', 'n_cells_by_counts', 'mean_counts', 'pct_dropout_by_counts', 'total_counts', 'ribo', 'hb', 'highly_variable', 'means', 'dispersions', 'dispersions_norm', 'mean', 'std'
uns: 'celltype_colors', 'hvg', 'leiden', 'leiden_colors', 'leiden_sizes', 'log1p', 'neighbors', 'paga', 'pca', 'rank_genes_groups', 'umap', 'aucell'
obsm: 'X_pca', 'X_umap', 'X_aucell'
varm: 'PCs'
obsp: 'connectivities', 'distances'
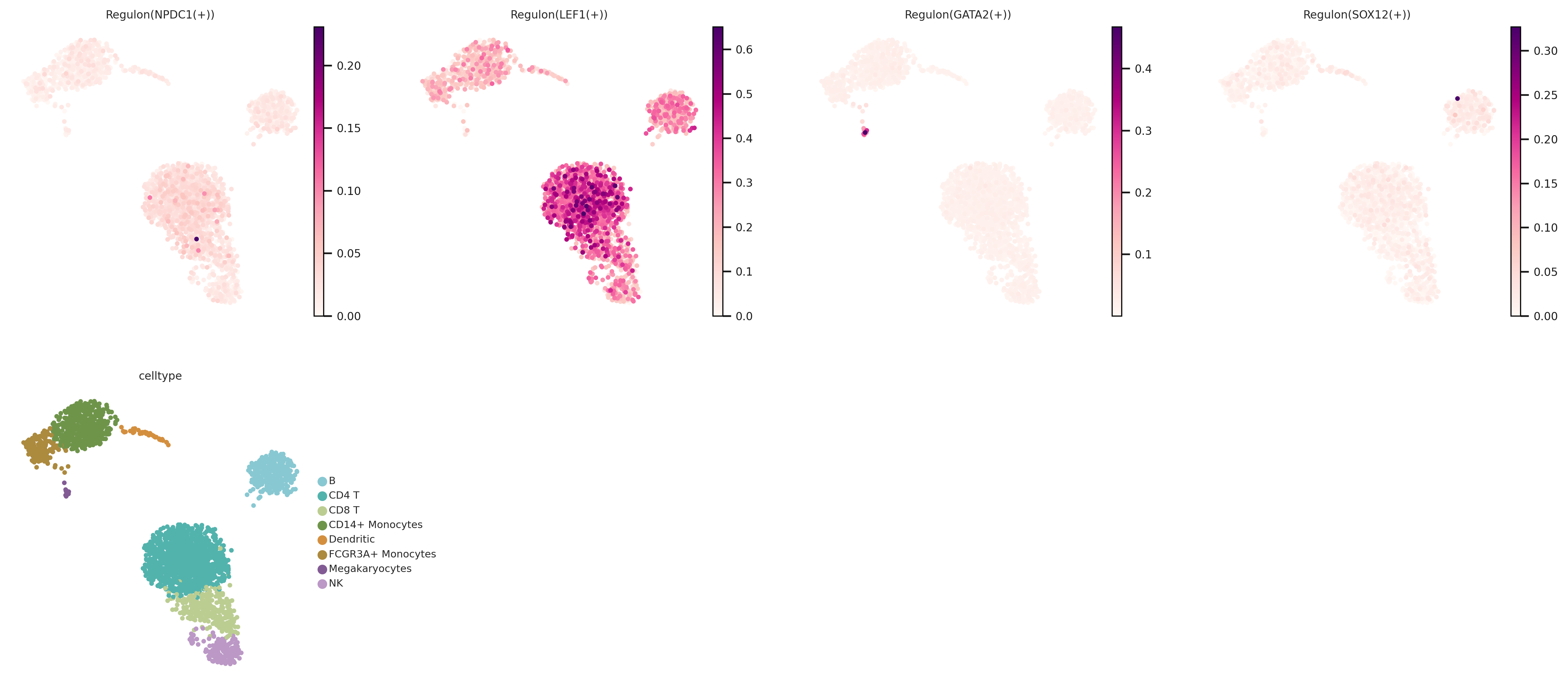
#选择需要呈现的转录因子进行展示
sc.set_figure_params(frameon=False, dpi=150, fontsize=8)
sc.pl.umap(pbmc, color=['Regulon(NPDC1(+))',
'Regulon(LEF1(+))',
'Regulon(GATA2(+))',
'Regulon(SOX12(+))','celltype'], ncols=4, cmap = "RdPu")
/home/zhaozm/anaconda3/envs/pyscenic/lib/python3.7/site-packages/IPython/core/pylabtools.py:151: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
fig.canvas.print_figure(bytes_io, **kw)
from matplotlib import cm, colors
from typing import Mapping, Sequence
from matplotlib.pyplot import rc_context
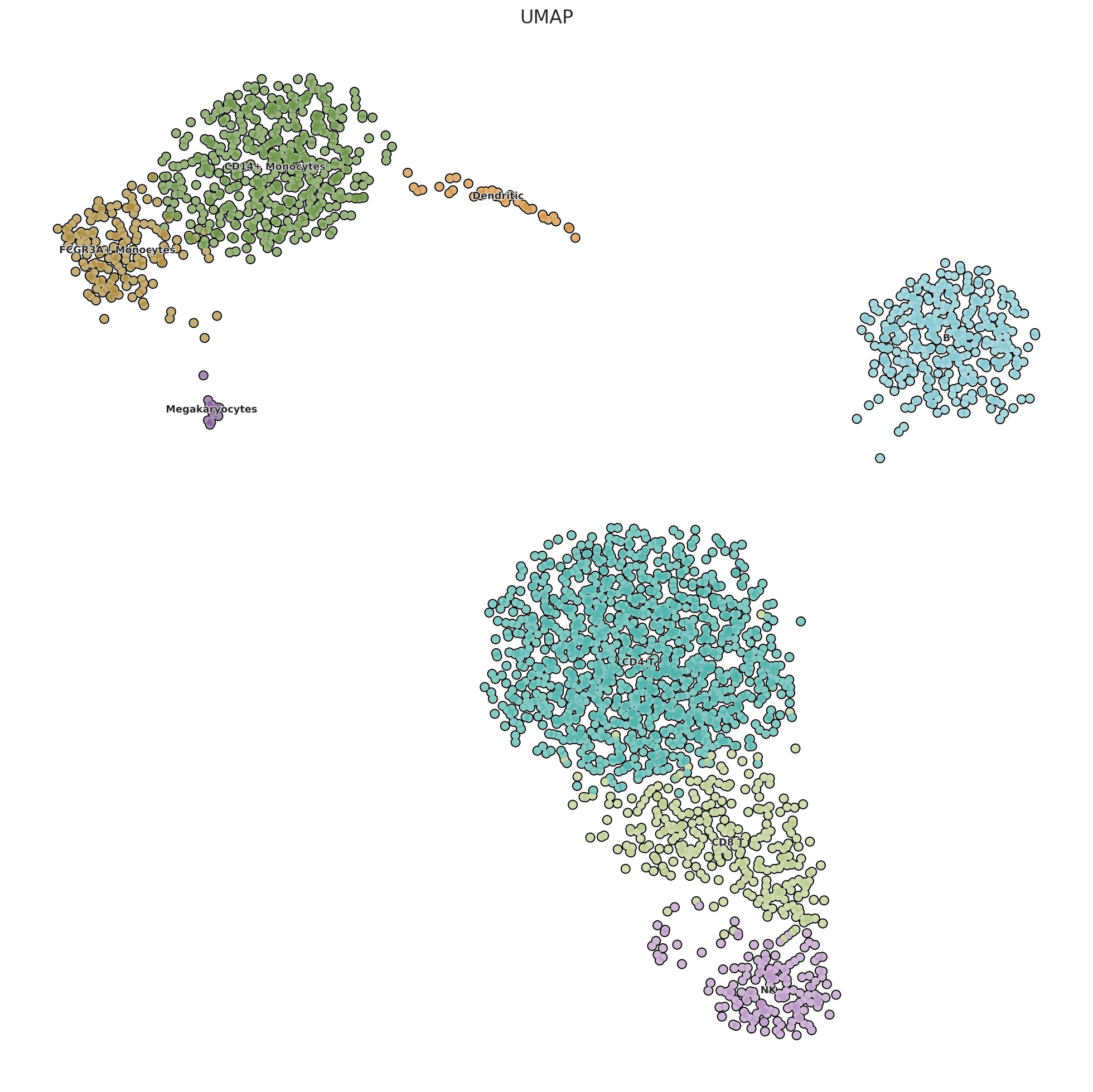
sc.set_figure_params(scanpy=True, fontsize=15)
with rc_context({'figure.figsize': (12.5, 12.5)}):
sc.pl.umap(pbmc, color='celltype',
add_outline=True, outline_width= [0.15,0.01],
legend_loc='on data',legend_fontsize=8, legend_fontoutline=1,
frameon=False,title='UMAP',size = 150)
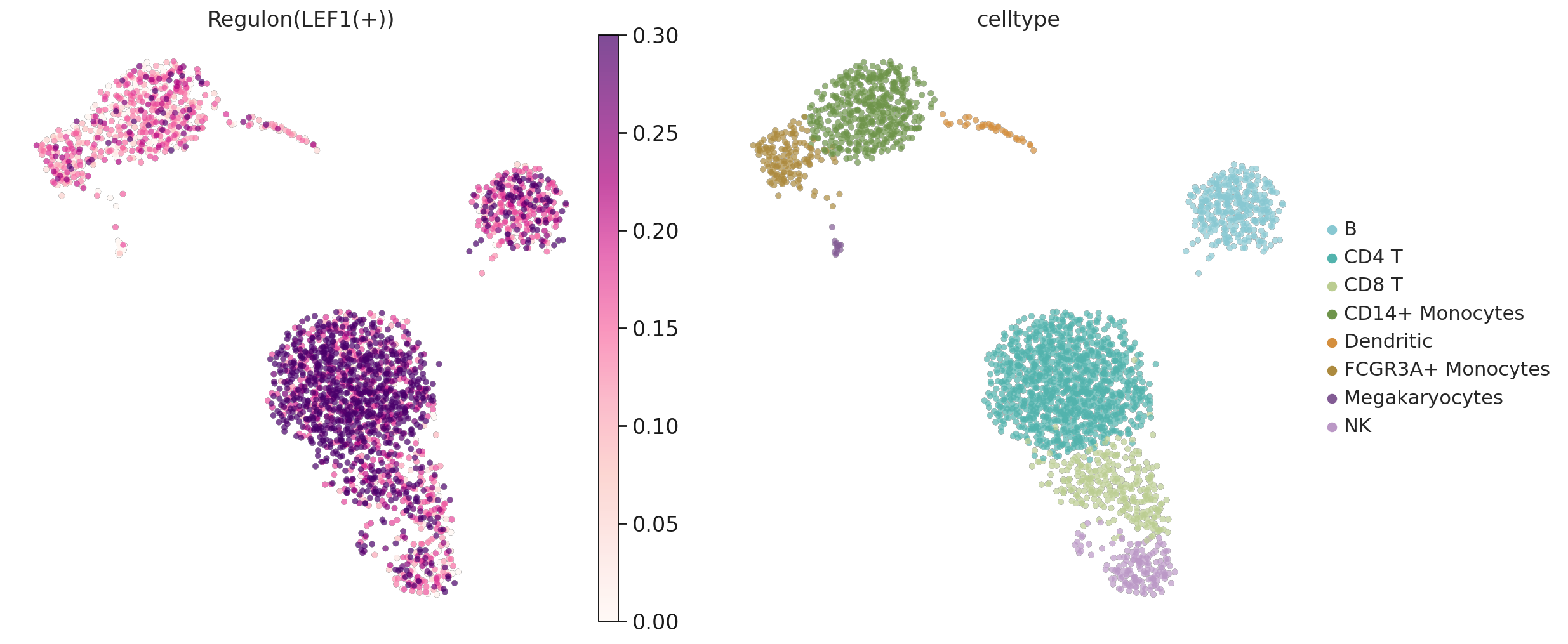
with rc_context({'figure.figsize': (7.5, 7.5)}):
sc.pl.umap(pbmc, color= ['Regulon(LEF1(+))', 'celltype'],
add_outline=True, outline_width= [0.01,0.01],
frameon=False,legend_fontweight='light',
cmap = "RdPu", size = 70, vmax= 0.3)
/home/zhaozm/anaconda3/envs/pyscenic/lib/python3.7/site-packages/IPython/core/pylabtools.py:151: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
fig.canvas.print_figure(bytes_io, **kw)